Cython#
Cython is a superset of Python#
Cython is a superset of Python, with additional functionality for defining C types and calling C functions
Cython generates C wrapper code, which is compiled into a Python extension module
Major advantage: enables incremental code optimization
Extensive documentation available on http://docs.cython.org
type annotations are used to declare C variables#
In this class, we are going to use Cython’s pure Python mode, which requires Cython 3.
and special handling of an module called cython
import cython as C
i: C.int
j: C.int
f: C.float
float_array: C.float[42]
float_ptr = C.pointer(C.float)
Cython also offers its own syntax in .pyx files.
Cython function definitions#
There are three kinds of Cython function definitions: def, cdef and cpdef:
# Python function (available to Python)
def foo(i: C.int, s: C.pointer(C.char):
...
# C function. Not visible to Python code that imports the module
@C.cfunc
def eggs(i: C.int, f: C.float) -> C.int:
...
# "Hybrid". Generates both Python and C functions.
@C.ccall
def foo_2(i: C.int, f: C.double) -> C.double:
...
Note: Function arguments and return types may be declared.
Cython optimises based on type definitions#
If no type is specified for a variable, parameter or return type, it defaults to a Python object
The standard Python for-loop is used in Cython:
i: C.int
n: C.int
for i in range(n):
...
If
iis declared as an integer (withi: C.int), this will be optimized into a standard C loop.
A Cython example#
Approximate the integral of a general function
f(x)
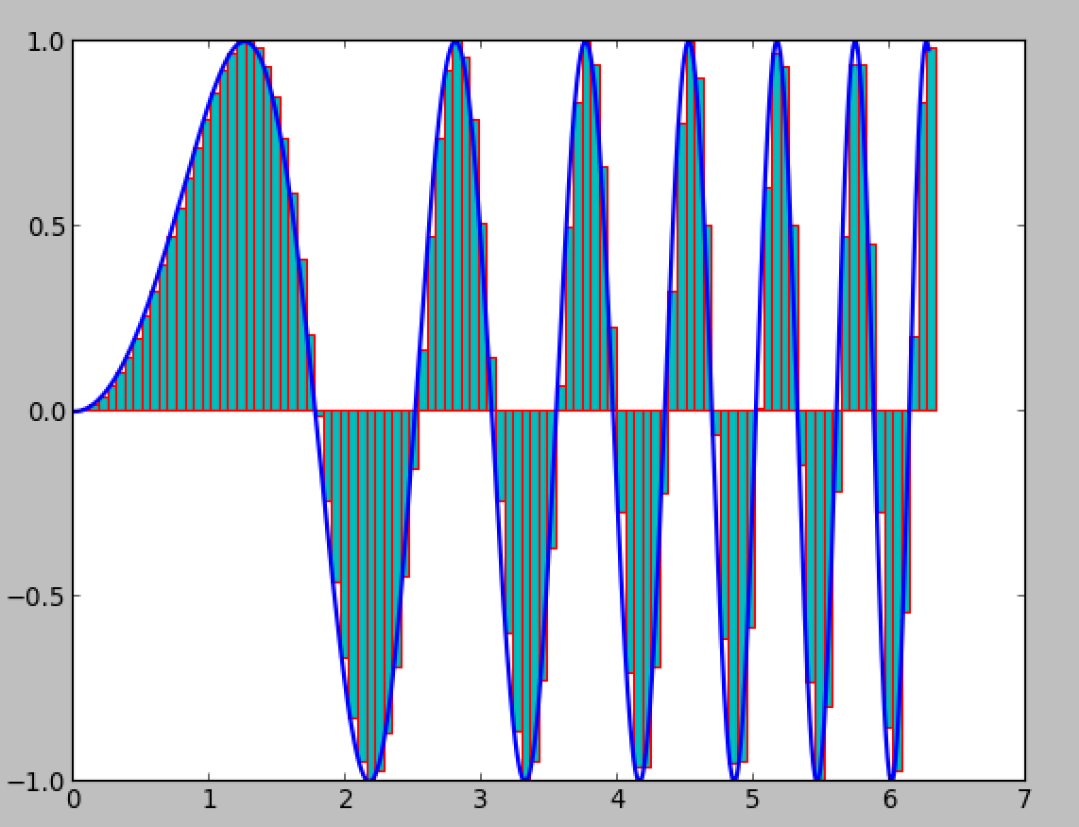
Numerical integration: accuracy increases with number of intervals
Speed is not a problem in 1D, but may be critical in 3D
Cython example: Standard Python#
Python implementation (not optimized) of the integration:
from math import sin
def f(x):
return sin(x**2)
def integrate_f(a, b, N):
s = 0
dx = (b - a) / N
for i in range(N):
s += f(a + i * dx)
return s * dx
N = 8_000_000
tr = %timeit -o integrate_f(0, 2, N)
993 ms ± 12.1 ms per loop (mean ± std. dev. of 7 runs, 1 loop each)
Integration takes around 1 second with N=8_000_000.
Cython example: Compilation with setuptools (recommended)#
Compiling with setuptools is more convenient.
Make a script named setup.py:
%pycat setup.py
from Cython.Build import cythonize
from setuptools import setup
setup(
name="in3110-cython",
ext_modules=cythonize(
["integral*.py", "apply.py"],
language_level=3,
annotate=True,
),
)
and compile the module with
!python3 setup.py build_ext --inplace
running build_ext
We can now import and run our compiled integral module
%pycat integral_notypes.py
from math import sin
def f(x):
return sin(x**2)
def integrate_f(a, b, N):
s = 0
dx = (b - a) / N
for i in range(N):
s += f(a + i * dx)
return s * dx
tr.average
0.9932964705720744
import integral_notypes
tr_notypes = %timeit -o integral_notypes.integrate_f(0, 2, N)
tr_notypes.average / tr.average
852 ms ± 17.8 ms per loop (mean ± std. dev. of 7 runs, 1 loop each)
0.8578035074871886
from timing_utils import timing_table
timings = {"python": tr, "cython (no types)": tr_notypes}
timing_table(
timings,
title="Cython example: Cython is only slightly faster than pure Python",
)
Cython example: Cython is only slightly faster than pure Python
| implementation | speed |
|---|---|
| python | 1.0 (normalized) |
| cython (no types) | 1.17x |
Cython example: adding types#
Simply compiling the Cython file gives only minor speedup: loop runs in C, but makes numerous calls to the Python/C API
To have any real speedup, we need to introduce types:
%pycat integral_types.py
from math import sin
import cython as C
def f(x: C.double) -> C.double:
return sin(x**2)
def integrate_f(a: C.double, b: C.double, N: C.int) -> C.double:
s: C.double = 0
dx: C.double = (b - a) / N
i: C.int
for i in range(N):
s += f(a + i * dx)
return s * dx
import integral_types
tr_types = %timeit -o integral_types.integrate_f(0, 2, N)
369 ms ± 6.59 ms per loop (mean ± std. dev. of 7 runs, 1 loop each)
timings["cython (types)"] = tr_types
timing_table(timings)
| implementation | speed |
|---|---|
| python | 1.0 (normalized) |
| cython (no types) | 1.17x |
| cython (types) | 2.69x |
Cython example: final version#
A fully typed version runs about 10 times faster:
from cython.cimports.libc.math import sin # Use cimport to make functions available to the C layer of Cython
@C.cfunc
def f(x: C.double) -> C.double:
return sin(x**2)
import integral
tr_cython = %timeit -o integral.integrate_f(0, 2, N)
23.9 ms ± 440 µs per loop (mean ± std. dev. of 7 runs, 10 loops each)
timings["cython (cfunc)"] = tr_cython
timing_table(
timings,
'Cython example: "less Python" equals "more speedup"',
)
Cython example: "less Python" equals "more speedup"
| implementation | speed |
|---|---|
| python | 1.0 (normalized) |
| cython (no types) | 1.17x |
| cython (types) | 2.69x |
| cython (cfunc) | 41.6x |
Speedup can be much higher, but requires slightly more complex example (loops within loops…).
You can also include your own C-functions, see https://cython.readthedocs.io/en/latest/src/tutorial/external.html.
Cython and numpy#
Cython works with numpy arrays as well.
Example: Apply sin to all numbers in an array:#
from math import sin
import numpy as np
def apply_sin(a):
out = np.empty_like(a)
for i in range(len(a)):
out[i] = sin(a[i])
return out
Usage:
a = np.linspace(0, 10, 1_000_000, dtype=np.double)
tr_sin = %timeit -o apply_sin(a)
99.2 ms ± 1.08 ms per loop (mean ± std. dev. of 7 runs, 10 loops each)
Declaring numpy data types#
Cython uses “typed memoryviews” for generating efficient C code for working with the data in numpy arrays. Below is the translation table between Python and Cython dypes:
from cython import int_types
Numpy datatype |
Cython datatype |
|---|---|
numpy.uint8 |
cython.cimports.libc.stdint.uint8_t |
numpy.int16 |
cython.cimports.libc.stdint.int16_t |
numpy.single |
cython.float |
numpy.double |
cython.double |
numpy.complex |
cython.complex |
Defining a new numpy array in Cython:
from cython import double
out: double[:]
out = numpy.zeros(1000, dtype=numpy.double)
Declaring numpy data types#
Below is a fully typed version of the apply_sin function:
%pycat apply.py
import numpy as np
from cython import double, int
from cython.cimports.libc.math import sin
def apply_sin(a: double[:]) -> double[:]:
i: int
out: double[:] = np.empty_like(a)
for i in range(len(a)):
out[i] = sin(a[i])
return out
Using the Cython memoryview API#
Save this file as apply.py. Once compiled, the cython module can be used as:
import apply
tr_cython = %timeit -o out = apply.apply_sin(a)
3.8 ms ± 200 µs per loop (mean ± std. dev. of 7 runs, 100 loops each)
And compare the result with the numpy builtin:
tr_numpy = %timeit -o np.sin(a)
3.64 ms ± 153 µs per loop (mean ± std. dev. of 7 runs, 100 loops each)
timing_table(
{
"math.sin": tr_sin,
"Cython": tr_cython,
"numpy": tr_numpy,
}
)
| implementation | speed |
|---|---|
| math.sin | 1.0 (normalized) |
| Cython | 26.1x |
| numpy | 27.3x |
Cython summary#
Cython pros and cons
[+] Allows incremental optimization, easy to access C libraries, generated C code more compact and readable than swig, active developer community, advanced and flexible
[+] Pure Python syntax (requires Cython 3.0)
[-] Less suitable than e.g. pybind11 for wrapping large libraries to Python modules, fully optimized code not as readable as Python
Should be considered (maybe as a first choice?) for mixing Python with C