Visualizing corona case data with altair#
Norway has stopped tracking and reporting data about the ongoing covid pandemic, but we can view data from 2020-2022.
import io
from functools import lru_cache
import altair as alt
import matplotlib.pyplot as plt
import pandas as pd
import requests
data_url = "https://raw.githubusercontent.com/thohan88/covid19-nor-data/HEAD/data"
@lru_cache()
def download_dataset(path):
"""Download a dataset from covid19-nor-data archive"""
url = f"{data_url}/{path}"
print(f"Downloading {url}")
r = requests.get(f"{data_url}/{path}")
r.raise_for_status()
print(f"Downloaded {len(r.content) // 1024}kB")
if path.endswith(".csv"):
read = pd.read_csv
elif path.endswith(".xlsx"):
read = pd.read_excel
return read(io.BytesIO(r.content), parse_dates=["date"])
all_cases = download_dataset("01_infected/msis/municipality_and_district.csv")
all_cases
Downloading https://raw.githubusercontent.com/thohan88/covid19-nor-data/HEAD/data/01_infected/msis/municipality_and_district.csv
Downloaded 31065kB
| date | date_time | kommune_bydel_no | kommune_bydel_name | bydel_no | bydel_name | kommune_no | kommune_name | fylke_no | fylke_name | population | cases | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 2020-03-26 | 2020-03-26T04:00:00Z | 30101 | Gamle Oslo | 30101.0 | Gamle Oslo | 301 | Oslo | 3 | Oslo | 58671 | 67 |
| 1 | 2020-03-26 | 2020-03-26T04:00:00Z | 30102 | Grünerløkka | 30102.0 | Grünerløkka | 301 | Oslo | 3 | Oslo | 62423 | 59 |
| 2 | 2020-03-26 | 2020-03-26T04:00:00Z | 30103 | Sagene | 30103.0 | Sagene | 301 | Oslo | 3 | Oslo | 45089 | 47 |
| 3 | 2020-03-26 | 2020-03-26T04:00:00Z | 30104 | St. Hanshaugen | 30104.0 | St. Hanshaugen | 301 | Oslo | 3 | Oslo | 38945 | 33 |
| 4 | 2020-03-26 | 2020-03-26T04:00:00Z | 30105 | Frogner | 30105.0 | Frogner | 301 | Oslo | 3 | Oslo | 59269 | 67 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 377250 | 2022-12-07 | 2022-12-07T04:00:00Z | 5442 | Nesseby | NaN | NaN | 5442 | Nesseby | 54 | Troms og Finnmark | 926 | 88 |
| 377251 | 2022-12-07 | 2022-12-07T04:00:00Z | 5443 | Båtsfjord | NaN | NaN | 5443 | Båtsfjord | 54 | Troms og Finnmark | 2221 | 447 |
| 377252 | 2022-12-07 | 2022-12-07T04:00:00Z | 5444 | Sør-Varanger | NaN | NaN | 5444 | Sør-Varanger | 54 | Troms og Finnmark | 10158 | 1601 |
| 377253 | 2022-12-07 | 2022-12-07T04:00:00Z | 9998 | Svalbard | NaN | NaN | 9998 | Svalbard | 99 | Svalbard | 0 | 0 |
| 377254 | 2022-12-07 | 2022-12-07T04:00:00Z | 9999 | Ukjent Kommune | NaN | NaN | 9999 | Ukjent Kommune | 99 | Ukjent Fylke | 0 | 19722 |
377255 rows × 12 columns
# aggregate data by fylke
cases = (
all_cases.groupby(["fylke_name", "date"])[["cases", "population"]]
.sum()
.reset_index()
).copy()
cases
| fylke_name | date | cases | population | |
|---|---|---|---|---|
| 0 | Agder | 2020-03-26 | 133 | 307231 |
| 1 | Agder | 2020-03-27 | 148 | 307231 |
| 2 | Agder | 2020-03-28 | 159 | 307231 |
| 3 | Agder | 2020-03-29 | 161 | 307231 |
| 4 | Agder | 2020-03-30 | 166 | 307231 |
| ... | ... | ... | ... | ... |
| 12800 | Viken | 2022-12-03 | 393395 | 1241165 |
| 12801 | Viken | 2022-12-04 | 393399 | 1241165 |
| 12802 | Viken | 2022-12-05 | 393410 | 1241165 |
| 12803 | Viken | 2022-12-06 | 393463 | 1241165 |
| 12804 | Viken | 2022-12-07 | 393518 | 1241165 |
12805 rows × 4 columns
# discard ukjent fylke where population 100k doesn't make sense
cases = cases[~cases.fylke_name.str.contains("Ukjent")].copy()
cases
| fylke_name | date | cases | population | |
|---|---|---|---|---|
| 0 | Agder | 2020-03-26 | 133 | 307231 |
| 1 | Agder | 2020-03-27 | 148 | 307231 |
| 2 | Agder | 2020-03-28 | 159 | 307231 |
| 3 | Agder | 2020-03-29 | 161 | 307231 |
| 4 | Agder | 2020-03-30 | 166 | 307231 |
| ... | ... | ... | ... | ... |
| 12800 | Viken | 2022-12-03 | 393395 | 1241165 |
| 12801 | Viken | 2022-12-04 | 393399 | 1241165 |
| 12802 | Viken | 2022-12-05 | 393410 | 1241165 |
| 12803 | Viken | 2022-12-06 | 393463 | 1241165 |
| 12804 | Viken | 2022-12-07 | 393518 | 1241165 |
11820 rows × 4 columns
# 'cases' is a cumulative sum
# reverse that to calculate the daily new case count
# 1. createe a new column
cases["daily cases"] = 0
# 2. populate it per fylke
for fylke in cases.fylke_name.unique():
mask = cases.fylke_name == fylke
fylke_cases = cases.loc[mask]
diff = fylke_cases.cases.diff()
# set first value from cases
diff.iloc[0] = fylke_cases.iloc[0].cases
cases.loc[fylke_cases.index, "daily cases"] = diff.astype(int)
cases
| fylke_name | date | cases | population | daily cases | |
|---|---|---|---|---|---|
| 0 | Agder | 2020-03-26 | 133 | 307231 | 133 |
| 1 | Agder | 2020-03-27 | 148 | 307231 | 15 |
| 2 | Agder | 2020-03-28 | 159 | 307231 | 11 |
| 3 | Agder | 2020-03-29 | 161 | 307231 | 2 |
| 4 | Agder | 2020-03-30 | 166 | 307231 | 5 |
| ... | ... | ... | ... | ... | ... |
| 12800 | Viken | 2022-12-03 | 393395 | 1241165 | 47 |
| 12801 | Viken | 2022-12-04 | 393399 | 1241165 | 4 |
| 12802 | Viken | 2022-12-05 | 393410 | 1241165 | 11 |
| 12803 | Viken | 2022-12-06 | 393463 | 1241165 | 53 |
| 12804 | Viken | 2022-12-07 | 393518 | 1241165 | 55 |
11820 rows × 5 columns
Add per-100,000 population column#
# per100k is "daily new cases per 100k population"
cases["per100k"] = cases["daily cases"] * 1e5 / (cases["population"] + 1)
cases
| fylke_name | date | cases | population | daily cases | per100k | |
|---|---|---|---|---|---|---|
| 0 | Agder | 2020-03-26 | 133 | 307231 | 133 | 43.289761 |
| 1 | Agder | 2020-03-27 | 148 | 307231 | 15 | 4.882304 |
| 2 | Agder | 2020-03-28 | 159 | 307231 | 11 | 3.580356 |
| 3 | Agder | 2020-03-29 | 161 | 307231 | 2 | 0.650974 |
| 4 | Agder | 2020-03-30 | 166 | 307231 | 5 | 1.627435 |
| ... | ... | ... | ... | ... | ... | ... |
| 12800 | Viken | 2022-12-03 | 393395 | 1241165 | 47 | 3.786762 |
| 12801 | Viken | 2022-12-04 | 393399 | 1241165 | 4 | 0.322278 |
| 12802 | Viken | 2022-12-05 | 393410 | 1241165 | 11 | 0.886263 |
| 12803 | Viken | 2022-12-06 | 393463 | 1241165 | 53 | 4.270178 |
| 12804 | Viken | 2022-12-07 | 393518 | 1241165 | 55 | 4.431317 |
11820 rows × 6 columns
Exercise#
plot cases per 1000 by fylke over time
latest_cases = cases[-5000:]
latest_cases.columns
Index(['fylke_name', 'date', 'cases', 'population', 'daily cases', 'per100k'], dtype='object')
subset = cases[cases.fylke_name.isin(["Oslo", "Viken"])]
daily = (
alt.Chart(subset)
.mark_point()
.encode(
x="date:T",
y="per100k",
color="fylke_name",
tooltip=[
"fylke_name",
"population",
"per100k",
"cases",
],
)
)
daily
weekly = (
alt.Chart(subset)
.mark_line()
.transform_window(
weekly_avg="mean(per100k)",
frame=[-3, 3],
)
.encode(
x="date:T",
y="weekly_avg:Q",
color="fylke_name",
tooltip=[
"fylke_name",
"population",
"per100k",
"cases",
"date",
],
)
)
weekly
weekly | daily
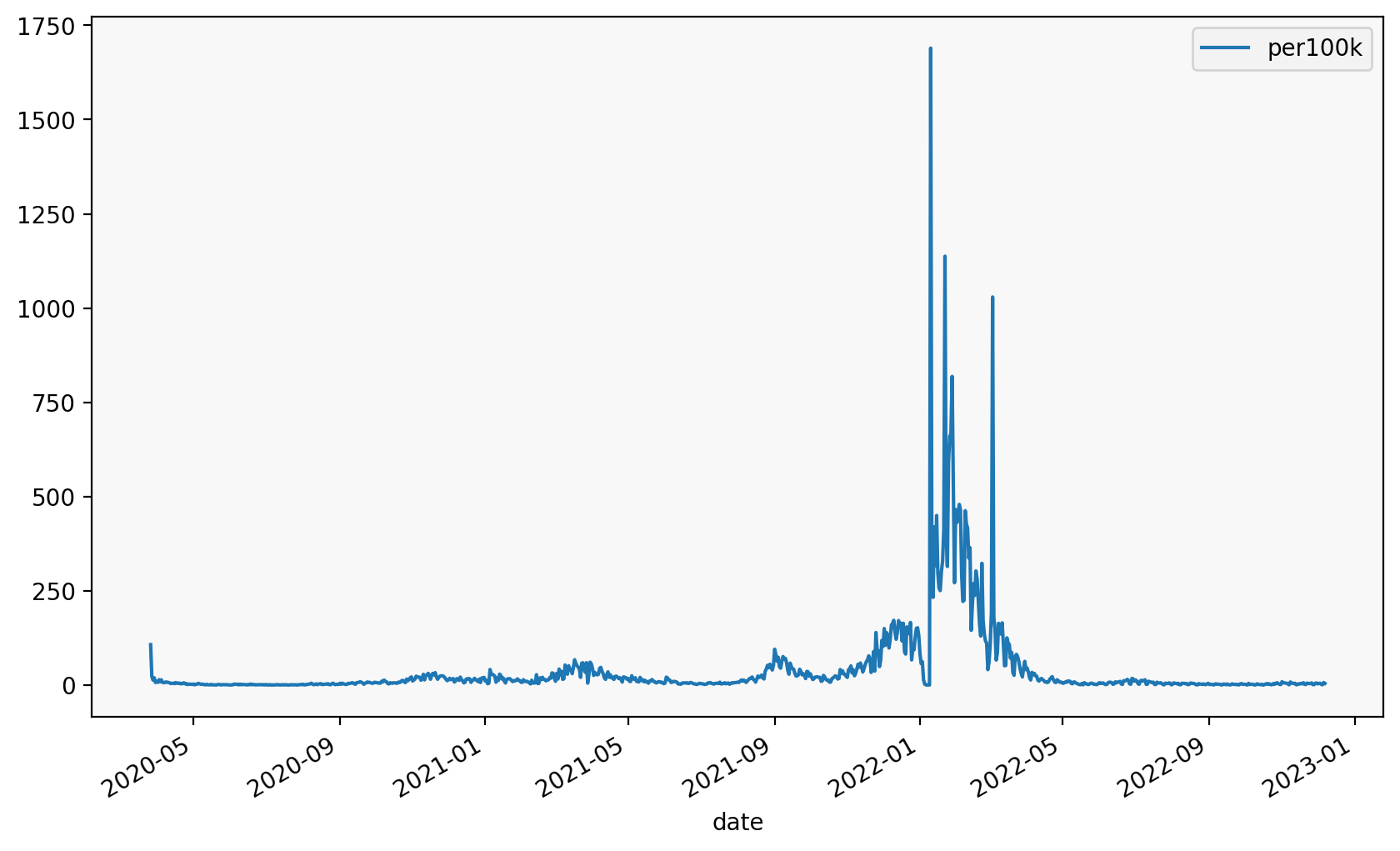
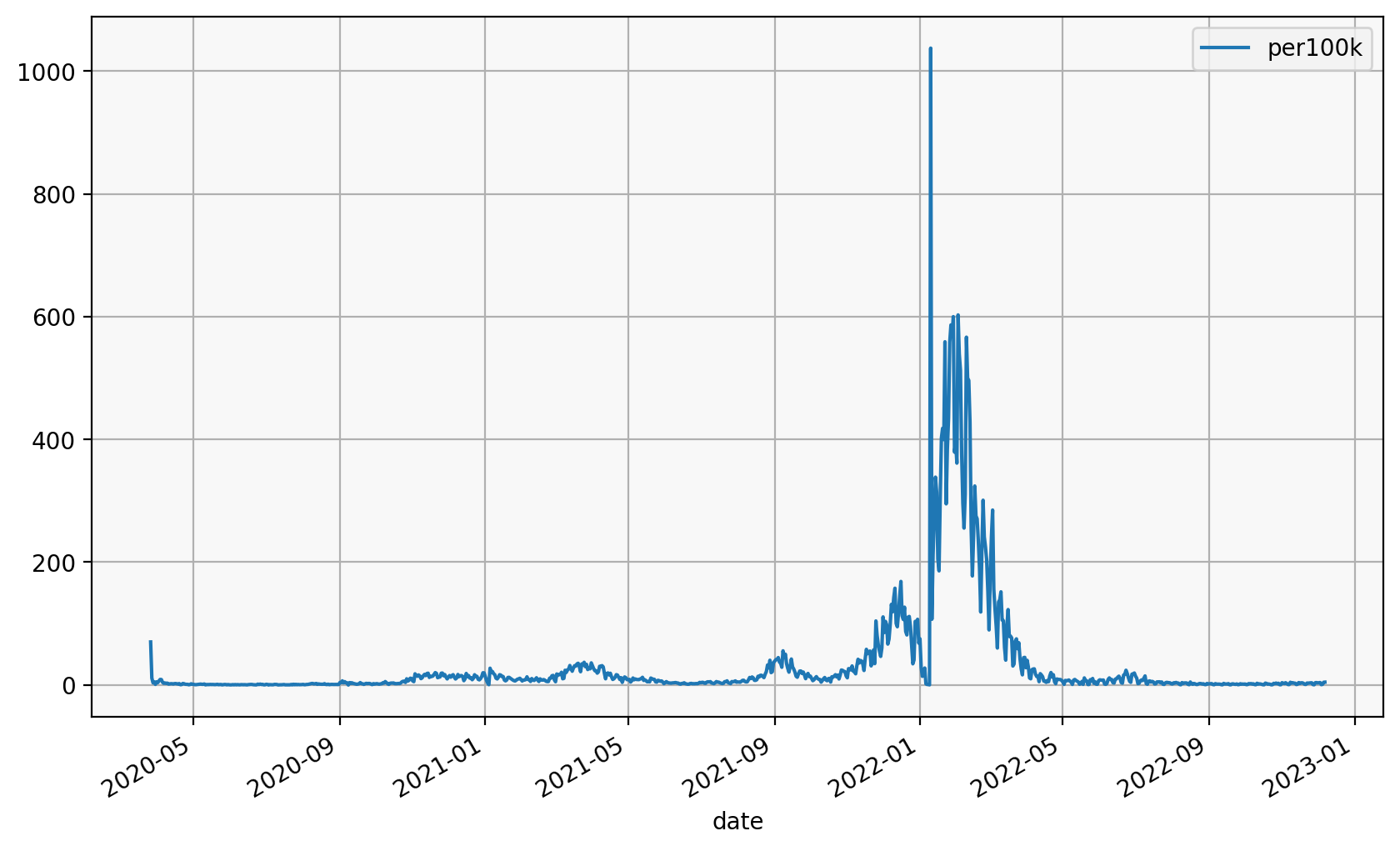
We can also do the same with matplotlib
subset.groupby("fylke_name")[["date", "per100k"]].plot(
x="date", y="per100k", legend=True
)
fylke_name
Oslo Axes(0.125,0.2;0.775x0.68)
Viken Axes(0.125,0.2;0.775x0.68)
dtype: object
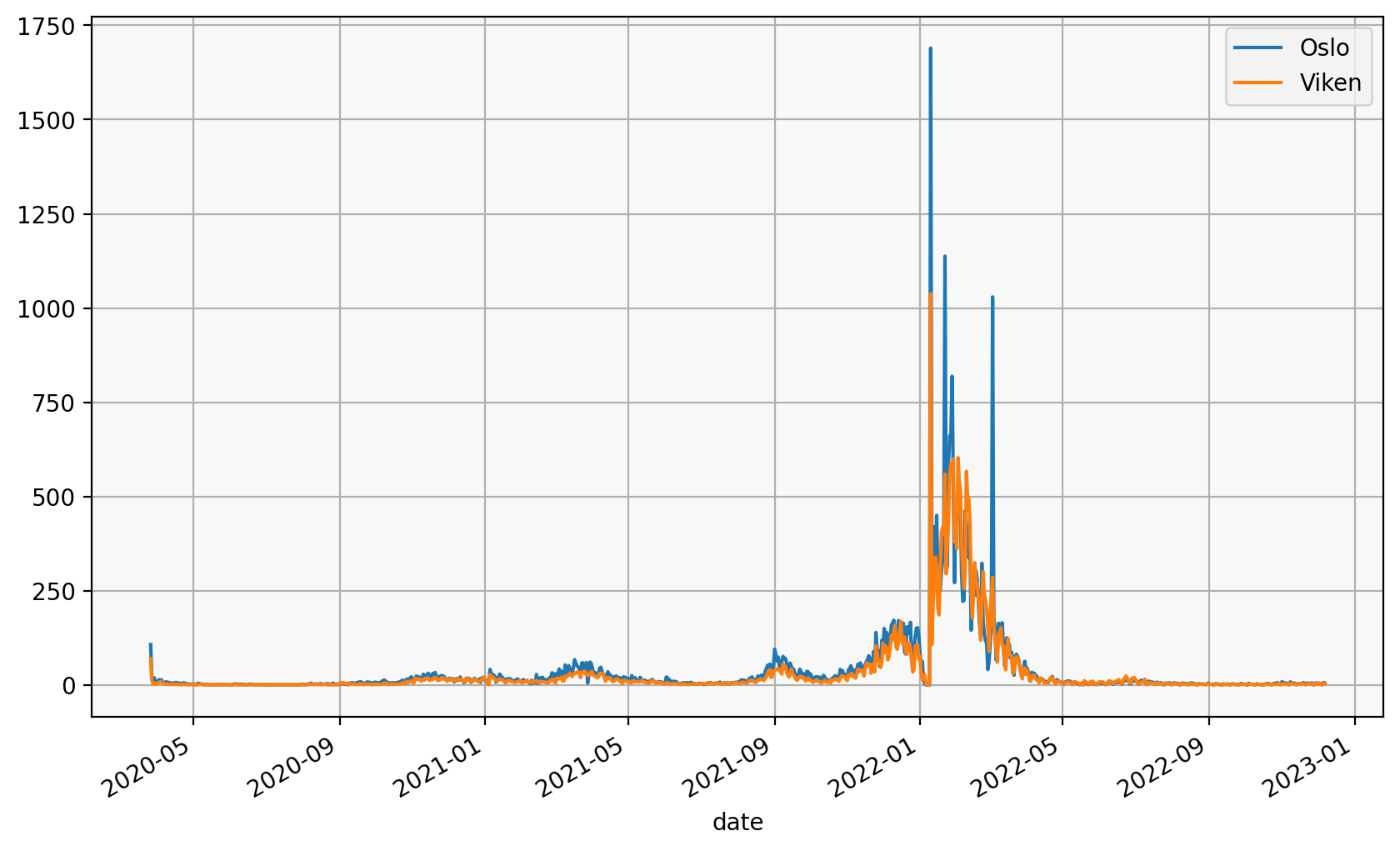
subset.set_index("date").groupby("fylke_name").per100k.plot(legend=True)
fylke_name
Oslo Axes(0.125,0.2;0.775x0.68)
Viken Axes(0.125,0.2;0.775x0.68)
Name: per100k, dtype: object
admissions = pd.read_csv(
"pasienter-innlagt-i-syke.csv",
parse_dates=["Dato"],
dayfirst=True,
)
admissions
| Dato | Kumulativt antall | Nye sykehusinnlegelser | |
|---|---|---|---|
| 0 | 2020-02-21 | 0 | 0 |
| 1 | 2020-02-22 | 0 | 0 |
| 2 | 2020-02-23 | 0 | 0 |
| 3 | 2020-02-24 | 0 | 0 |
| 4 | 2020-02-25 | 0 | 0 |
| ... | ... | ... | ... |
| 973 | 2022-10-21 | 16348 | 13 |
| 974 | 2022-10-22 | 16359 | 11 |
| 975 | 2022-10-23 | 16374 | 15 |
| 976 | 2022-10-24 | 16389 | 15 |
| 977 | 2022-10-25 | 16390 | 1 |
978 rows × 3 columns
alt.Chart(admissions).mark_line().encode(
x="Dato",
y="Nye sykehusinnlegelser:Q",
)
monthly_hospital = (
alt.Chart(admissions)
.mark_line()
.transform_window(
frame=[-30, 0],
admissions="mean(Nye sykehusinnlegelser)",
)
.encode(
x="Dato",
y="admissions:Q",
)
)
alt.vconcat(monthly_hospital, weekly)
weekly