Pandas - Data Analysis with Python#
(last update 11/10/23)
Pandas is a high-performance, easy-to-use data structures and data analysis tools.

There is abundance of data and we should/need to make sense of it
Smart devices, Strava, …
SSB Statistics Norway *
EuroStat Statistics Europe *
Kaggle Data Science compatitions*
Quandl for finances
(We will work with these ones today in lecture* or exercises**)
What is Pandas good for?#
Working with (large) data sets and created automated data processes.
Pandas is extensively used to prepare data in data science (machine learning, data analytics, …)
Examples:
Import and export data into standard formats (CSV, Excel, Latex, ..).
Combine with Numpy for advanced computations or Matplotlib for visualisations.
Calculate statistics and answer questions about the data, like
What’s the average, median, max, or min of each column?
Does column A correlate with column B?
What does the distribution of data in column C look like?
Clean up data (e.g. fill out missing information and fix inconsistent formatting) and merge multiple data sets into one common one
More information#
Official Pandas documentation: http://pandas.pydata.org/pandas-docs/stable/tutorials.html
Pandas cookbook: http://pandas.pydata.org/pandas-docs/stable/cookbook.html
Wes McKinney, Python for Data Analysis

Python Data Science Handbook by Jake VanderPlas (We follow Chapter 4 in this lecture)

This lecture#
Part1 Introduction to Pandas
Part2 Hands on examples with real data
Installation#
If you have Anaconda: Already installed
If you have Miniconda:
conda install pandasIf you have your another Python distribution:
python3 -m pip install pandas
Let’s dive in
import pandas as pd
Pandas Series object#
Series is 1d series of data similar to numpy.array.
series = pd.Series([4, 5, 6, 7, 8])
series.values
array([4, 5, 6, 7, 8])
series.index
RangeIndex(start=0, stop=5, step=1)
(series.values.dtype, series.index.dtype)
(dtype('int64'), dtype('int64'))
We see that series are indexed and both values and index are typed. As we saw with numpy this has performance benefits. The similarity with numpy.array
series[0]
4
import numpy as np
np.power(series, 2)
0 16
1 25
2 36
3 49
4 64
dtype: int64
However, the indices do not need to be numbers (and neither need to be the values)
values = list(range(65, 75))
index = [chr(v) for v in values]
series = pd.Series(values, index=index)
series
A 65
B 66
C 67
D 68
E 69
F 70
G 71
H 72
I 73
J 74
dtype: int64
The series than behaves also like a dictionary, although it supports fancy indexing.
series["A"], series["D":"H"]
(65,
D 68
E 69
F 70
G 71
H 72
dtype: int64)
Continuing the analogy with dictionary a possible way to make series is from a dictionary
data = {
"Washington": "United States of America",
"London": "Great Britain",
"Oslo": "Norway",
}
series = pd.Series(data)
series
Washington United States of America
London Great Britain
Oslo Norway
dtype: object
Note that for values that are amenable to str we have the string methods …
series.str.upper()
Washington UNITED STATES OF AMERICA
London GREAT BRITAIN
Oslo NORWAY
dtype: object
… and so we can for example compute a mask using regular expressions (here looking for states with 2 word names) …
mask = series.str.match(r"^\w+\s+\w+$")
mask
Washington False
London True
Oslo False
dtype: bool
… which can be used to index into the series
series[mask]
London Great Britain
dtype: object
Some other examples of series indexing
(series[0], series["London":"Oslo"], series[0:2])
('United States of America',
London Great Britain
Oslo Norway
dtype: object,
Washington United States of America
London Great Britain
dtype: object)
We can be more explicit about the indexing with indexers loc and iloc
(series.iloc[0], series.loc["London":"Oslo"], series.iloc[0:2])
('United States of America',
London Great Britain
Oslo Norway
dtype: object,
Washington United States of America
London Great Britain
dtype: object)
series[1:3]
London Great Britain
Oslo Norway
dtype: object
import this
The Zen of Python, by Tim Peters
Beautiful is better than ugly.
Explicit is better than implicit.
Simple is better than complex.
Complex is better than complicated.
Flat is better than nested.
Sparse is better than dense.
Readability counts.
Special cases aren't special enough to break the rules.
Although practicality beats purity.
Errors should never pass silently.
Unless explicitly silenced.
In the face of ambiguity, refuse the temptation to guess.
There should be one-- and preferably only one --obvious way to do it.
Although that way may not be obvious at first unless you're Dutch.
Now is better than never.
Although never is often better than *right* now.
If the implementation is hard to explain, it's a bad idea.
If the implementation is easy to explain, it may be a good idea.
Namespaces are one honking great idea -- let's do more of those!
But why does this matter? Consider
tricky = pd.Series(["a", "b", "c"], index=[7, 5, 4])
tricky
7 a
5 b
4 c
dtype: object
# Let's demonstrate that we can sort
tricky = tricky.sort_index()
tricky
4 c
5 b
7 a
dtype: object
# Here we are refering to the value at row where index = 4 and values where at first though second row!
# Try tricky.loc[4] vs tricky.iloc[4]
(tricky.loc[4], tricky[0:2])
('c',
4 c
5 b
dtype: object)
Pandas DataFrame object#
Two dimensional data are represented by DataFrames. Again Pandas allows for flexibility of what row/columns indices can be. DataFrames can be constructed in a number of ways
def spreadsheet(nrows, ncols, start=0, cstart=0, base=0):
data = base + np.random.rand(nrows, ncols)
index = [f"r{i}" for i in range(start, start + nrows)]
columns = [f"c{i}" for i in range(cstart, cstart + ncols)]
# c0 c1 c2
# r0
# r1
return pd.DataFrame(data, index=index, columns=columns)
data_frame = spreadsheet(10, 4)
data_frame["c0"]
r0 0.341584
r1 0.154689
r2 0.829218
r3 0.860563
r4 0.482496
r5 0.623338
r6 0.951465
r7 0.330926
r8 0.805647
r9 0.038046
Name: c0, dtype: float64
For large frame it is usefull to look at portions of the data
data_frame = spreadsheet(10_000, 4)
print(len(data_frame))
data_frame.head(10) # data_frame.tail is for the end part
10000
| c0 | c1 | c2 | c3 | |
|---|---|---|---|---|
| r0 | 0.658119 | 0.888360 | 0.151696 | 0.680974 |
| r1 | 0.519551 | 0.838755 | 0.180593 | 0.933276 |
| r2 | 0.149844 | 0.110505 | 0.253935 | 0.506687 |
| r3 | 0.582279 | 0.171603 | 0.414131 | 0.822162 |
| r4 | 0.721373 | 0.428403 | 0.404248 | 0.841991 |
| r5 | 0.432620 | 0.953647 | 0.023574 | 0.614961 |
| r6 | 0.939906 | 0.018228 | 0.556107 | 0.012206 |
| r7 | 0.836547 | 0.463066 | 0.802324 | 0.835970 |
| r8 | 0.440520 | 0.777486 | 0.042400 | 0.008710 |
| r9 | 0.165533 | 0.267046 | 0.287619 | 0.101397 |
We can combines multiple series into a frame
index = ("Min", "Ingeborg", "Miro")
nationality = pd.Series(["USA", "NOR", "SVK"], index=index)
university = pd.Series(["Berkeley", "Bergen", "Oslo"], index=index)
instructors = pd.DataFrame({"nat": nationality, "uni": university})
instructors
| nat | uni | |
|---|---|---|
| Min | USA | Berkeley |
| Ingeborg | NOR | Bergen |
| Miro | SVK | Oslo |
(instructors.index, instructors.columns)
(Index(['Min', 'Ingeborg', 'Miro'], dtype='object'),
Index(['nat', 'uni'], dtype='object'))
Anothor way of creating is using dictionaries
f = pd.DataFrame(
{
"country": ["CZE", "NOR", "USA"],
"capital": ["Prague", "Oslo", "Washington DC"],
"pop": [5, 10, 300],
}
)
f
| country | capital | pop | |
|---|---|---|---|
| 0 | CZE | Prague | 5 |
| 1 | NOR | Oslo | 10 |
| 2 | USA | Washington DC | 300 |
When some column is unique we can designate it as index. Of course, tables can be stored in various formats. We will come back to reading later.
# LaTex
import re
fi = f.set_index("country")
tex_table = fi.style.to_latex()
# To ease the prining
for row in re.split(r"\n", tex_table):
print(row)
\begin{tabular}{llr}
& capital & pop \\
country & & \\
CZE & Prague & 5 \\
NOR & Oslo & 10 \\
USA & Washington DC & 300 \\
\end{tabular}
# CSV
tex_table = fi.to_csv(sep=";")
# To ease the prining
for row in re.split(r"\n", tex_table):
print(row)
country;capital;pop
CZE;Prague;5
NOR;Oslo;10
USA;Washington DC;300
Going back to frame creation, recall that above, the two series had a common index. What happens with the frame when this is not the case? Below we also illustrate another constructor.
nationality = {"Miro": "SVK", "Ingeborg": "NOR", "Min": "USA"}
university = {"Miro": "Oslo", "Ingeborg": "Bergen", "Min": "Berkeley", "Joe": "Harvard"}
office = {"Miro": 303, "Ingeborg": 309, "Min": 311, "Joe": -1}
# Construct from list of dictionary using union of keys as columns - we need to fill in values for some
# NOTE: here each dictionary essentially defines a row
missing_data_frame = pd.DataFrame(
[nationality, university, office], index=["nation", "uni", "office"]
)
missing_data_frame
| Miro | Ingeborg | Min | Joe | |
|---|---|---|---|---|
| nation | SVK | NOR | USA | NaN |
| uni | Oslo | Bergen | Berkeley | Harvard |
| office | 303 | 309 | 311 | -1 |
# Just to make it look better
missing_data_frame = missing_data_frame.T
missing_data_frame
| nation | uni | office | |
|---|---|---|---|
| Miro | SVK | Oslo | 303 |
| Ingeborg | NOR | Bergen | 309 |
| Min | USA | Berkeley | 311 |
| Joe | NaN | Harvard | -1 |
The missing value has been filled with a special value. Real data often suffer from lack of regularity. We can check for this
missing_data_frame.isna()
| nation | uni | office | |
|---|---|---|---|
| Miro | False | False | False |
| Ingeborg | False | False | False |
| Min | False | False | False |
| Joe | True | False | False |
And the provide a missing value. Note that this can be done already when we are constructing the frame.
missing_data_frame.fillna("OutThere")
| nation | uni | office | |
|---|---|---|---|
| Miro | SVK | Oslo | 303 |
| Ingeborg | NOR | Bergen | 309 |
| Min | USA | Berkeley | 311 |
| Joe | OutThere | Harvard | -1 |
Or discard the data. We can drop the row which has NaN. Note that by default any NaN is enough but we can be more tolerant (see how and thresh keyword arguments)
missing_data_frame.dropna()
| nation | uni | office | |
|---|---|---|---|
| Miro | SVK | Oslo | 303 |
| Ingeborg | NOR | Bergen | 309 |
| Min | USA | Berkeley | 311 |
Or we drop the problematic column. Axis 0 is row (just like in numpy)
missing_data_frame.dropna(axis=1)
| uni | office | |
|---|---|---|
| Miro | Oslo | 303 |
| Ingeborg | Bergen | 309 |
| Min | Berkeley | 311 |
| Joe | Harvard | -1 |
Note that the above calls are returning a new Frame, ie. are not in in-place. For this use .dropna(..., inplace=True)
Once we have the frame we can start computing with it. Let’s consider indexing first
missing_data_frame
| nation | uni | office | |
|---|---|---|---|
| Miro | SVK | Oslo | 303 |
| Ingeborg | NOR | Bergen | 309 |
| Min | USA | Berkeley | 311 |
| Joe | NaN | Harvard | -1 |
# Get a specific series
missing_data_frame["nation"]
Miro SVK
Ingeborg NOR
Min USA
Joe NaN
Name: nation, dtype: object
# Or several as a frame
missing_data_frame[["nation", "uni"]]
| nation | uni | |
|---|---|---|
| Miro | SVK | Oslo |
| Ingeborg | NOR | Bergen |
| Min | USA | Berkeley |
| Joe | NaN | Harvard |
Note that slicing default to rows
# Will fail because no rows names like that
missing_data_frame["nation":"office"]
---------------------------------------------------------------------------
KeyError Traceback (most recent call last)
File ~/.local/lib/python3.9/site-packages/pandas/core/indexes/base.py:3653, in Index.get_loc(self, key)
3652 try:
-> 3653 return self._engine.get_loc(casted_key)
3654 except KeyError as err:
File ~/.local/lib/python3.9/site-packages/pandas/_libs/index.pyx:147, in pandas._libs.index.IndexEngine.get_loc()
File ~/.local/lib/python3.9/site-packages/pandas/_libs/index.pyx:176, in pandas._libs.index.IndexEngine.get_loc()
File pandas/_libs/hashtable_class_helper.pxi:7080, in pandas._libs.hashtable.PyObjectHashTable.get_item()
File pandas/_libs/hashtable_class_helper.pxi:7088, in pandas._libs.hashtable.PyObjectHashTable.get_item()
KeyError: 'nation'
The above exception was the direct cause of the following exception:
KeyError Traceback (most recent call last)
Cell In [37], line 2
1 # Will fail because no rows names like that
----> 2 missing_data_frame["nation":"office"]
File ~/.local/lib/python3.9/site-packages/pandas/core/frame.py:3735, in DataFrame.__getitem__(self, key)
3733 # Do we have a slicer (on rows)?
3734 if isinstance(key, slice):
-> 3735 indexer = self.index._convert_slice_indexer(key, kind="getitem")
3736 if isinstance(indexer, np.ndarray):
3737 # reachable with DatetimeIndex
3738 indexer = lib.maybe_indices_to_slice(
3739 indexer.astype(np.intp, copy=False), len(self)
3740 )
File ~/.local/lib/python3.9/site-packages/pandas/core/indexes/base.py:4132, in Index._convert_slice_indexer(self, key, kind)
4130 indexer = key
4131 else:
-> 4132 indexer = self.slice_indexer(start, stop, step)
4134 return indexer
File ~/.local/lib/python3.9/site-packages/pandas/core/indexes/base.py:6344, in Index.slice_indexer(self, start, end, step)
6300 def slice_indexer(
6301 self,
6302 start: Hashable | None = None,
6303 end: Hashable | None = None,
6304 step: int | None = None,
6305 ) -> slice:
6306 """
6307 Compute the slice indexer for input labels and step.
6308
(...)
6342 slice(1, 3, None)
6343 """
-> 6344 start_slice, end_slice = self.slice_locs(start, end, step=step)
6346 # return a slice
6347 if not is_scalar(start_slice):
File ~/.local/lib/python3.9/site-packages/pandas/core/indexes/base.py:6537, in Index.slice_locs(self, start, end, step)
6535 start_slice = None
6536 if start is not None:
-> 6537 start_slice = self.get_slice_bound(start, "left")
6538 if start_slice is None:
6539 start_slice = 0
File ~/.local/lib/python3.9/site-packages/pandas/core/indexes/base.py:6462, in Index.get_slice_bound(self, label, side)
6459 return self._searchsorted_monotonic(label, side)
6460 except ValueError:
6461 # raise the original KeyError
-> 6462 raise err
6464 if isinstance(slc, np.ndarray):
6465 # get_loc may return a boolean array, which
6466 # is OK as long as they are representable by a slice.
6467 assert is_bool_dtype(slc.dtype)
File ~/.local/lib/python3.9/site-packages/pandas/core/indexes/base.py:6456, in Index.get_slice_bound(self, label, side)
6454 # we need to look up the label
6455 try:
-> 6456 slc = self.get_loc(label)
6457 except KeyError as err:
6458 try:
File ~/.local/lib/python3.9/site-packages/pandas/core/indexes/base.py:3655, in Index.get_loc(self, key)
3653 return self._engine.get_loc(casted_key)
3654 except KeyError as err:
-> 3655 raise KeyError(key) from err
3656 except TypeError:
3657 # If we have a listlike key, _check_indexing_error will raise
3658 # InvalidIndexError. Otherwise we fall through and re-raise
3659 # the TypeError.
3660 self._check_indexing_error(key)
KeyError: 'nation'
# See that slice goes for rows
missing_data_frame["Miro":"Min"]
| nation | uni | office | |
|---|---|---|---|
| Miro | SVK | Oslo | 303 |
| Ingeborg | NOR | Bergen | 309 |
| Min | USA | Berkeley | 311 |
We can call indexers to rescue
missing_data_frame.loc[:, "nation":"uni"]
| nation | uni | |
|---|---|---|
| Miro | SVK | Oslo |
| Ingeborg | NOR | Bergen |
| Min | USA | Berkeley |
| Joe | NaN | Harvard |
Indexes can of course be more involed
missing_data_frame["office"] > 305
Miro False
Ingeborg True
Min True
Joe False
Name: office, dtype: bool
missing_data_frame.loc[missing_data_frame["office"] > 305, ["uni", "nation"]]
| uni | nation | |
|---|---|---|
| Ingeborg | Bergen | NOR |
| Min | Berkeley | USA |
Next let’s compute with the values in the frames
f0 = spreadsheet(4, 4)
np.cos(f0)
| c0 | c1 | c2 | c3 | |
|---|---|---|---|---|
| r0 | 0.870461 | 0.789699 | 0.739024 | 0.860132 |
| r1 | 0.785803 | 0.718719 | 0.580812 | 0.584109 |
| r2 | 0.988713 | 0.956762 | 0.956623 | 0.851564 |
| r3 | 0.559086 | 0.889881 | 0.990609 | 0.903777 |
Binary operation will align the frame - operation is valid only for the index-column pairs found in both and with will get missing for the rest
f1 = spreadsheet(5, 6)
f0 + f1
| c0 | c1 | c2 | c3 | c4 | c5 | |
|---|---|---|---|---|---|---|
| r0 | 0.533858 | 0.997285 | 1.098653 | 1.032666 | NaN | NaN |
| r1 | 1.372231 | 0.804163 | 1.445381 | 1.393992 | NaN | NaN |
| r2 | 1.056678 | 0.809150 | 0.589316 | 0.936123 | NaN | NaN |
| r3 | 1.311115 | 0.863243 | 0.404915 | 0.961394 | NaN | NaN |
| r4 | NaN | NaN | NaN | NaN | NaN | NaN |
Note that this is note unique to frames and works (unsuprisingly on series too)
c00 = f0["c0"]
c10 = f1["c0"]
c00 + c10
r0 0.533858
r1 1.372231
r2 1.056678
r3 1.311115
r4 NaN
Name: c0, dtype: float64
Reduction operators default to be computed for each column
f1.max()
c0 0.906288
c1 0.514013
c2 0.883873
c3 0.519110
c4 0.910645
c5 0.811385
dtype: float64
For column wise reduction we need to specify the axis (axis=1)
f1.mean(axis="columns")
r0 0.339750
r1 0.554993
r2 0.498857
r3 0.482604
r4 0.406889
dtype: float64
Pandas Index object#
In the previous usecase f0 + f1 we have seen that operation is performed for common keys - this suggests that we can do logical operations on indices. This is indeed the case
i0 = f0.index
i1 = f1.index
(i0.union(i1), i0.intersection(i1))
(Index(['r0', 'r1', 'r2', 'r3', 'r4'], dtype='object'),
Index(['r0', 'r1', 'r2', 'r3'], dtype='object'))
Indexes can be explicitely constructed. For working with time series it is useful to index by time. Let’s get timestamp indexing for every Day between 2 dates
import datetime
time = pd.date_range(
start=datetime.date.fromisoformat("2022-10-19"),
end=datetime.date.fromisoformat("2022-12-20"),
freq="D",
)
time
DatetimeIndex(['2022-10-19', '2022-10-20', '2022-10-21', '2022-10-22',
'2022-10-23', '2022-10-24', '2022-10-25', '2022-10-26',
'2022-10-27', '2022-10-28', '2022-10-29', '2022-10-30',
'2022-10-31', '2022-11-01', '2022-11-02', '2022-11-03',
'2022-11-04', '2022-11-05', '2022-11-06', '2022-11-07',
'2022-11-08', '2022-11-09', '2022-11-10', '2022-11-11',
'2022-11-12', '2022-11-13', '2022-11-14', '2022-11-15',
'2022-11-16', '2022-11-17', '2022-11-18', '2022-11-19',
'2022-11-20', '2022-11-21', '2022-11-22', '2022-11-23',
'2022-11-24', '2022-11-25', '2022-11-26', '2022-11-27',
'2022-11-28', '2022-11-29', '2022-11-30', '2022-12-01',
'2022-12-02', '2022-12-03', '2022-12-04', '2022-12-05',
'2022-12-06', '2022-12-07', '2022-12-08', '2022-12-09',
'2022-12-10', '2022-12-11', '2022-12-12', '2022-12-13',
'2022-12-14', '2022-12-15', '2022-12-16', '2022-12-17',
'2022-12-18', '2022-12-19', '2022-12-20'],
dtype='datetime64[ns]', freq='D')
We generate the corresponding data and illustrate some plotting capabilities of Pandas
import matplotlib.pyplot as plt
values = np.sin(np.pi / 10 * np.arange(len(time)))
time_series = pd.Series(values, index=time)
time_series.plot()
<AxesSubplot: >
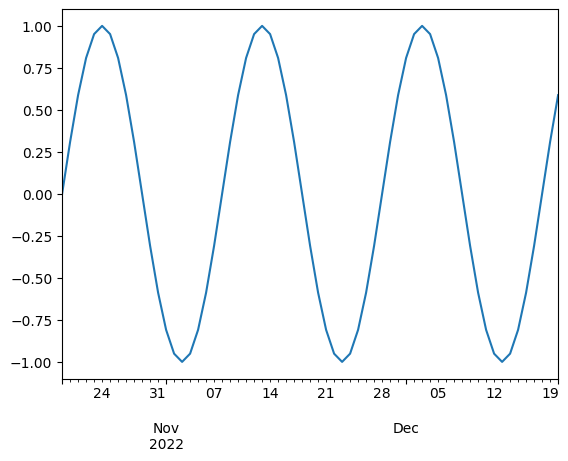
Note that we are getting the nice xlabels for free!
Time indexing allows us to do some fancy opearations. For example we can have a look at the signal for only the first day of the week (Monday?). For other functionality see resample, ‘rolling’ means on windows ets.
time_series[time.weekday == 0].plot()
<AxesSubplot: >
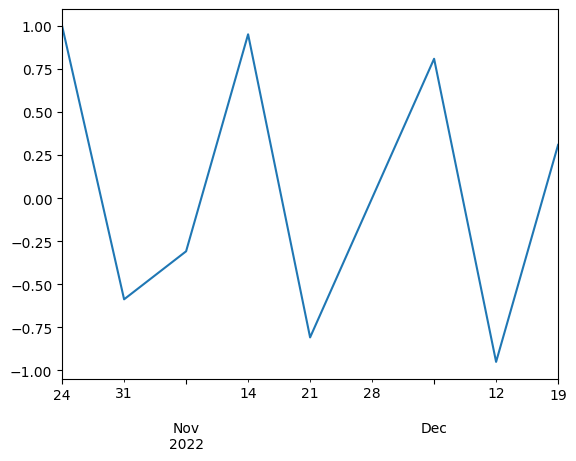
Note the value of difference between to time indices
time[:4] - time[1:5]
TimedeltaIndex(['-1 days', '-1 days', '-1 days', '-1 days'], dtype='timedelta64[ns]', freq=None)
The Timedelta object can be useful for shifting
diff = pd.Timedelta("1m")
time = time + diff
time[:5]
DatetimeIndex(['2022-10-19 00:01:00', '2022-10-20 00:01:00',
'2022-10-21 00:01:00', '2022-10-22 00:01:00',
'2022-10-23 00:01:00'],
dtype='datetime64[ns]', freq='D')
Time stamps often come in different format. Pandas offers convenience functions for their conversion/parsing.
df = pd.DataFrame({"date": [1470195805, 1480195805, 1490195805], "value": [2, 3, 4]})
df
| date | value | |
|---|---|---|
| 0 | 1470195805 | 2 |
| 1 | 1480195805 | 3 |
| 2 | 1490195805 | 4 |
When was the clock started?
pd.to_datetime(df["date"], unit="s")
0 2016-08-03 03:43:25
1 2016-11-26 21:30:05
2 2017-03-22 15:16:45
Name: date, dtype: datetime64[ns]
import time
pd.to_datetime(pd.Series([0, time.time()]), unit="s")
0 1970-01-01 00:00:00.000000000
1 2023-10-12 07:10:21.943605760
dtype: datetime64[ns]
Question: how can we get the years prior to 1970 to work?
Hierarchical indexing with MultiIndex#
Multiindexing is a way to handle higher order tensors, e.g. f(x, y, color). It also allows for organizing the data by establishing hierarchy in indexing. Suppose we have RGB image of 5 x 4 pixels. We could represent this as a table with channel x and y coordinate columns and 5 x 4 x 3 rows. A representation more true to the nature of the data would be to have a “column” for each color
column_index = pd.MultiIndex.from_product(
[["R", "G", "B"], [0, 1, 2, 3]], names=("color", "cindex")
)
column_index
MultiIndex([('R', 0),
('R', 1),
('R', 2),
('R', 3),
('G', 0),
('G', 1),
('G', 2),
('G', 3),
('B', 0),
('B', 1),
('B', 2),
('B', 3)],
names=['color', 'cindex'])
image = pd.DataFrame(
np.arange(60).reshape(5, 12),
index=pd.Index([0, 1, 2, 3, 4], name="rindex"),
columns=column_index,
)
image
| color | R | G | B | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| cindex | 0 | 1 | 2 | 3 | 0 | 1 | 2 | 3 | 0 | 1 | 2 | 3 |
| rindex | ||||||||||||
| 0 | 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 |
| 1 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | 23 |
| 2 | 24 | 25 | 26 | 27 | 28 | 29 | 30 | 31 | 32 | 33 | 34 | 35 |
| 3 | 36 | 37 | 38 | 39 | 40 | 41 | 42 | 43 | 44 | 45 | 46 | 47 |
| 4 | 48 | 49 | 50 | 51 | 52 | 53 | 54 | 55 | 56 | 57 | 58 | 59 |
We can have a look at the said “flat” representation of the data
image.unstack()
color cindex rindex
R 0 0 0
1 12
2 24
3 36
4 48
1 0 1
1 13
2 25
3 37
4 49
2 0 2
1 14
2 26
3 38
4 50
3 0 3
1 15
2 27
3 39
4 51
G 0 0 4
1 16
2 28
3 40
4 52
1 0 5
1 17
2 29
3 41
4 53
2 0 6
1 18
2 30
3 42
4 54
3 0 7
1 19
2 31
3 43
4 55
B 0 0 8
1 20
2 32
3 44
4 56
1 0 9
1 21
2 33
3 45
4 57
2 0 10
1 22
2 34
3 46
4 58
3 0 11
1 23
2 35
3 47
4 59
dtype: int64
Accessing the entries works as follows
image["R"] # Column(s) where first multiindex is R
image.loc[:, ("R", 0)] # Column multiindex by R, 0
image.loc[2, ("R", 1)] # The entry
25
We can perform reduction operation
image.mean().mean()
29.5
Combining datasets - appending#
Extend numpy.concatenate to pandas objects. We can combine data from 2 frames if they have some “axes” in common. Let’s start with 2 identical columns and do default vertical/row wise concatenation. Note that all the operations below create new DataFrame
# Unique row indices
d1 = spreadsheet(2, 2, base=0)
d2 = spreadsheet(4, 2, base=10)
print(d1)
print(d2)
c0 c1
r0 0.904955 0.461721
r1 0.728287 0.227259
c0 c1
r0 10.784765 10.696760
r1 10.949264 10.610865
r2 10.835922 10.139623
r3 10.458615 10.799789
pd.concat([d1, d2])
| c0 | c1 | |
|---|---|---|
| r0 | 0.904955 | 0.461721 |
| r1 | 0.728287 | 0.227259 |
| r0 | 10.784765 | 10.696760 |
| r1 | 10.949264 | 10.610865 |
| r2 | 10.835922 | 10.139623 |
| r3 | 10.458615 | 10.799789 |
What if there are duplicate indices?
d1 = spreadsheet(2, 2, base=0) # Has r0, r1
d2 = spreadsheet(2, 2, base=10) # Has r0, r1
print(d1)
print(d2)
d12 = pd.concat([d1, d2])
d12
c0 c1
r0 0.887823 0.685323
r1 0.952741 0.543635
c0 c1
r0 10.876731 10.797502
r1 10.161474 10.832729
| c0 | c1 | |
|---|---|---|
| r0 | 0.887823 | 0.685323 |
| r1 | 0.952741 | 0.543635 |
| r0 | 10.876731 | 10.797502 |
| r1 | 10.161474 | 10.832729 |
We can still get values but this lack of uniqueness might not be desirable and we might want to check for it…
d12.loc["r0", :]
| c0 | c1 | |
|---|---|---|
| r0 | 0.887823 | 0.685323 |
| r0 | 10.876731 | 10.797502 |
pd.concat([d1, d2], verify_integrity=False) # True
| c0 | c1 | |
|---|---|---|
| r0 | 0.887823 | 0.685323 |
| r1 | 0.952741 | 0.543635 |
| r0 | 10.876731 | 10.797502 |
| r1 | 10.161474 | 10.832729 |
… or reset the index.
pd.concat([d1, d2], ignore_index=True)
| c0 | c1 | |
|---|---|---|
| 0 | 0.887823 | 0.685323 |
| 1 | 0.952741 | 0.543635 |
| 2 | 10.876731 | 10.797502 |
| 3 | 10.161474 | 10.832729 |
A different, more organized option is to introduce multiindex to remember where the data came from
d12 = pd.concat([d1, d2], keys=["D1", "D2"])
d12
| c0 | c1 | ||
|---|---|---|---|
| D1 | r0 | 0.887823 | 0.685323 |
| r1 | 0.952741 | 0.543635 | |
| D2 | r0 | 10.876731 | 10.797502 |
| r1 | 10.161474 | 10.832729 |
d12.loc["D1", :]
| c0 | c1 | |
|---|---|---|
| r0 | 0.887823 | 0.685323 |
| r1 | 0.952741 | 0.543635 |
For frames sharing row index we can vertical/column wise concatenation
d1 = spreadsheet(2, 3, cstart=0)
d2 = spreadsheet(2, 3, cstart=3, start=5)
print(d1)
print(d2)
pd.concat([d1, d2], axis=1, verify_integrity=True)
c0 c1 c2
r0 0.022688 0.677420 0.705416
r1 0.877029 0.949243 0.317691
c3 c4 c5
r5 0.492914 0.206868 0.292487
r6 0.824141 0.039968 0.829352
| c0 | c1 | c2 | c3 | c4 | c5 | |
|---|---|---|---|---|---|---|
| r0 | 0.022688 | 0.677420 | 0.705416 | NaN | NaN | NaN |
| r1 | 0.877029 | 0.949243 | 0.317691 | NaN | NaN | NaN |
| r5 | NaN | NaN | NaN | 0.492914 | 0.206868 | 0.292487 |
| r6 | NaN | NaN | NaN | 0.824141 | 0.039968 | 0.829352 |
d1 = spreadsheet(2, 3, cstart=0)
d2 = spreadsheet(2, 3, cstart=3, start=1)
print(d1)
print(d2)
pd.concat([d1, d2], axis=1, verify_integrity=True)
c0 c1 c2
r0 0.178587 0.155051 0.513482
r1 0.540436 0.585551 0.965622
c3 c4 c5
r1 0.782119 0.806388 0.859816
r2 0.574038 0.615266 0.536074
| c0 | c1 | c2 | c3 | c4 | c5 | |
|---|---|---|---|---|---|---|
| r0 | 0.178587 | 0.155051 | 0.513482 | NaN | NaN | NaN |
| r1 | 0.540436 | 0.585551 | 0.965622 | 0.782119 | 0.806388 | 0.859816 |
| r2 | NaN | NaN | NaN | 0.574038 | 0.615266 | 0.536074 |
What if we are appending two tables with slighlty different columns. It makes sense that there would be some missing data
d1 = spreadsheet(2, 2, cstart=0) # Has c0 c1
d2 = spreadsheet(2, 3, start=0, cstart=1) # c1 c2 c3
print(d1)
print(d2)
pd.concat([d1, d2], ignore_index=True)
c0 c1
r0 0.151458 0.059844
r1 0.623653 0.035702
c1 c2 c3
r0 0.946713 0.906110 0.300365
r1 0.514724 0.687296 0.905855
| c0 | c1 | c2 | c3 | |
|---|---|---|---|---|
| 0 | 0.151458 | 0.059844 | NaN | NaN |
| 1 | 0.623653 | 0.035702 | NaN | NaN |
| 2 | NaN | 0.946713 | 0.906110 | 0.300365 |
| 3 | NaN | 0.514724 | 0.687296 | 0.905855 |
We see that the by default we combine columns from both tables. But we can be more specific using the join or join_axes keyword arguments
pd.concat([d1, d2], ignore_index=True, join="inner") # Use the column intersection c1
| c1 | |
|---|---|
| 0 | 0.059844 |
| 1 | 0.035702 |
| 2 | 0.946713 |
| 3 | 0.514724 |
Combining datasets - use relations to extend the data#
For pd.concant a typical use case is if we have tables of records (e.g. some logs of [transaction id, card number], [transaction id, amount]) and we want to append them. A different of way of building up/joining data is by pd.merge. Consider the following “relational tables”. From nationality we know that “Min” -> “USA” while at the same time university maps “Min” to “Berkeley”.
nationality = pd.DataFrame(
{
"name": ["Min", "Miro", "Ingeborg", "Aslak"],
"nation": ["USA", "SVK", "NOR", "NOR"],
}
)
print(nationality)
university = pd.DataFrame(
{
"name": ["Miro", "Ingeborg", "Min", "Aslak"],
"uni": ["Oslo", "Bergen", "Berkeley", "Oslo"],
}
)
print(university)
name nation
0 Min USA
1 Miro SVK
2 Ingeborg NOR
3 Aslak NOR
name uni
0 Miro Oslo
1 Ingeborg Bergen
2 Min Berkeley
3 Aslak Oslo
Note that the frame are not aligned in their indexes but using pd.merge we can still capture the connection between “Min”, “Berkely” and “USA”
pd.merge(nationality, university)
| name | nation | uni | |
|---|---|---|---|
| 0 | Min | USA | Berkeley |
| 1 | Miro | SVK | Oslo |
| 2 | Ingeborg | NOR | Bergen |
| 3 | Aslak | NOR | Oslo |
Above is an example of 1-1 join. We can go further and infer properties. Let’s have a look up for capitals of some states.
capitals = pd.DataFrame(
{"nation": ["USA", "SVK", "NOR"], "city": ["Washington", "Brasislava", "Oslo"]}
)
capitals
| nation | city | |
|---|---|---|
| 0 | USA | Washington |
| 1 | SVK | Brasislava |
| 2 | NOR | Oslo |
Combining nationality with capitals we can see that “Ingeborg -> NOR -> Oslo” and thus can build a table with cities
pd.merge(nationality, capitals)
| name | nation | city | |
|---|---|---|---|
| 0 | Min | USA | Washington |
| 1 | Miro | SVK | Brasislava |
| 2 | Ingeborg | NOR | Oslo |
| 3 | Aslak | NOR | Oslo |
Above the merge happened on unique common columns. Sometimes we want/need to be more explicit about the “relation”.
# This table does not have the "nation" column
capitals2 = pd.DataFrame(
{
"state": ["USA", "SVK", "NOR"],
"city": ["Washington", "Brasislava", "Oslo"],
"count": [4, 5, 6],
}
)
capitals2
| state | city | count | |
|---|---|---|---|
| 0 | USA | Washington | 4 |
| 1 | SVK | Brasislava | 5 |
| 2 | NOR | Oslo | 6 |
We can specify which columns are to be used to build the relation as follows
# We will have nation and state but they represent the same so drop
pd.merge(nationality, capitals2, left_on="nation", right_on="state").drop(
"nation", axis="columns"
)
| name | state | city | count | |
|---|---|---|---|---|
| 0 | Min | USA | Washington | 4 |
| 1 | Miro | SVK | Brasislava | 5 |
| 2 | Ingeborg | NOR | Oslo | 6 |
| 3 | Aslak | NOR | Oslo | 6 |
Indexes for row can also be used to this end. Let’s make a new frame which use state column as index.
capitals2
| state | city | count | |
|---|---|---|---|
| 0 | USA | Washington | 4 |
| 1 | SVK | Brasislava | 5 |
| 2 | NOR | Oslo | 6 |
capitals_ri = capitals2.set_index("state")
capitals_ri
| city | count | |
|---|---|---|
| state | ||
| USA | Washington | 4 |
| SVK | Brasislava | 5 |
| NOR | Oslo | 6 |
It’s merge can now be done in two ways. The old one …
pd.merge(nationality, capitals_ri, left_on="nation", right_on="state")
| name | nation | city | count | |
|---|---|---|---|---|
| 0 | Min | USA | Washington | 4 |
| 1 | Miro | SVK | Brasislava | 5 |
| 2 | Ingeborg | NOR | Oslo | 6 |
| 3 | Aslak | NOR | Oslo | 6 |
… or using row indexing
pd.merge(nationality, capitals_ri, left_on="nation", right_index=True)
| name | nation | city | count | |
|---|---|---|---|---|
| 0 | Min | USA | Washington | 4 |
| 1 | Miro | SVK | Brasislava | 5 |
| 2 | Ingeborg | NOR | Oslo | 6 |
| 3 | Aslak | NOR | Oslo | 6 |
What should happen if the relation cannot be establised for all the values needed? In the example below, we cannot infer position for all the names
in the nationality frame. A sensible default is to perform merge only for those cases where it is well defined.
jobs = pd.DataFrame({"who": ["Ingeborg", "Aslak"], "position": ["researcher", "ceo"]})
pd.merge(nationality, jobs, left_on="name", right_on="who")
| name | nation | who | position | |
|---|---|---|---|---|
| 0 | Ingeborg | NOR | Ingeborg | researcher |
| 1 | Aslak | NOR | Aslak | ceo |
However, by specifying the how keyword we can proceed with NaN for “outer” all the merged values or those in “left” or “right” frame.
pd.merge(nationality, jobs, left_on="name", right_on="who", how="outer") # Inner
| name | nation | who | position | |
|---|---|---|---|---|
| 0 | Min | USA | NaN | NaN |
| 1 | Miro | SVK | NaN | NaN |
| 2 | Ingeborg | NOR | Ingeborg | researcher |
| 3 | Aslak | NOR | Aslak | ceo |
Finally, pd.merge can yield to duplicate column indices. These are automatically made unique by suffixes
pd.merge(jobs, jobs, on="who", suffixes=["_left", "_right"])
| who | position_left | position_right | |
|---|---|---|---|
| 0 | Ingeborg | researcher | researcher |
| 1 | Aslak | ceo | ceo |
Data aggregation and transformations#
We have previously seen that we can extact information about min, max, mean etc where by default reduction happend for each fixed column. Hierchical indexing was one option to get some structure to what was being reduced. Here we introduce groupby which serves this purpose too.
data = pd.DataFrame(
{
"brand": ["tesla", "tesla", "tesla", "vw", "vw", "mercedes"],
"model": ["S", "3", "X", "tuareg", "passat", "Gclass"],
"type": ["sedan", "sedan", "suv", "suv", "sedan", "suv"],
"hp": [32, 43, 54, 102, 30, 50],
}
)
data
| brand | model | type | hp | |
|---|---|---|---|---|
| 0 | tesla | S | sedan | 32 |
| 1 | tesla | 3 | sedan | 43 |
| 2 | tesla | X | suv | 54 |
| 3 | vw | tuareg | suv | 102 |
| 4 | vw | passat | sedan | 30 |
| 5 | mercedes | Gclass | suv | 50 |
Let us begin by “fixing” a table - say we want to capitalize the “brand” data, e.g. mercedes -> Mercedes. Half-way solution could be
data["brand"] = data["brand"].str.upper()
data
| brand | model | type | hp | |
|---|---|---|---|---|
| 0 | TESLA | S | sedan | 32 |
| 1 | TESLA | 3 | sedan | 43 |
| 2 | TESLA | X | suv | 54 |
| 3 | VW | tuareg | suv | 102 |
| 4 | VW | passat | sedan | 30 |
| 5 | MERCEDES | Gclass | suv | 50 |
A better way is to use transform function to modify one series
data["brand"].transform(str.capitalize)
0 Tesla
1 Tesla
2 Tesla
3 Vw
4 Vw
5 Mercedes
Name: brand, dtype: object
Update the table by assignment
data["brand"] = data["brand"].transform(str.capitalize)
data
| brand | model | type | hp | |
|---|---|---|---|---|
| 0 | Tesla | S | sedan | 32 |
| 1 | Tesla | 3 | sedan | 43 |
| 2 | Tesla | X | suv | 54 |
| 3 | Vw | tuareg | suv | 102 |
| 4 | Vw | passat | sedan | 30 |
| 5 | Mercedes | Gclass | suv | 50 |
transform mapping can be specified not only via functions but also strings (which map to numpy)
data["hp"].transform("sqrt")
0 5.656854
1 6.557439
2 7.348469
3 10.099505
4 5.477226
5 7.071068
Name: hp, dtype: float64
data["hp"].transform(["sqrt", "cos"]) # Get several series out
| sqrt | cos | |
|---|---|---|
| 0 | 5.656854 | 0.834223 |
| 1 | 6.557439 | 0.555113 |
| 2 | 7.348469 | -0.829310 |
| 3 | 10.099505 | 0.101586 |
| 4 | 5.477226 | 0.154251 |
| 5 | 7.071068 | 0.964966 |
When modifying the frame we can specify which series is specified by which function
data.transform({"hp": "sqrt", "type": lambda x: len(x)})
| hp | type | |
|---|---|---|
| 0 | 5.656854 | 5 |
| 1 | 6.557439 | 5 |
| 2 | 7.348469 | 3 |
| 3 | 10.099505 | 3 |
| 4 | 5.477226 | 5 |
| 5 | 7.071068 | 3 |
Note that simply passing len would not work. Therefore we pass in lambda and the transformation is applied element by element. In general, this way of computing will be less performant - Python’s for loop vs. (numpy) vectorization
Finally, we look at apply of the table. Here we get access to the entire row, i.e. we can compute with multiplire series.
def foo(x):
x["w"] = x["hp"] * 0.7
return x
data.apply(foo, axis="columns") # NOTE: the new column index!
| brand | model | type | hp | w | |
|---|---|---|---|---|---|
| 0 | Tesla | S | sedan | 32 | 22.4 |
| 1 | Tesla | 3 | sedan | 43 | 30.1 |
| 2 | Tesla | X | suv | 54 | 37.8 |
| 3 | Vw | tuareg | suv | 102 | 71.4 |
| 4 | Vw | passat | sedan | 30 | 21.0 |
| 5 | Mercedes | Gclass | suv | 50 | 35.0 |
Now that we are happy with the table we now want to gather information based on brand. If we split by brand we can think of 3 (as many as unique brand values) subtables. Within these subtables we perform further operations/reductions - they values with be combined in the final result.
g = data.groupby("brand", group_keys=True)
g
<pandas.core.groupby.generic.DataFrameGroupBy object at 0x7f8a4aa25f10>
Note that we do not get a frame back but instead there is a DataFrameGroupBy object. This way we do not compute rightaway which could be expensive. We can verify that it it’s behavior is similar to the frame we wanted
g.indices
{'Mercedes': array([5]), 'Tesla': array([0, 1, 2]), 'Vw': array([3, 4])}
The object supports many methods of the DataFrame so we can continue “defining” our processing pipiline. For example we can get the column
g["hp"]
<pandas.core.groupby.generic.SeriesGroupBy object at 0x7f8a4b4dae50>
And finally force the computation
g["hp"].max()
brand
Mercedes 50
Tesla 54
Vw 102
Name: hp, dtype: int64
More complete description by describe
g["hp"].describe()
| count | mean | std | min | 25% | 50% | 75% | max | |
|---|---|---|---|---|---|---|---|---|
| brand | ||||||||
| Mercedes | 1.0 | 50.0 | NaN | 50.0 | 50.0 | 50.0 | 50.0 | 50.0 |
| Tesla | 3.0 | 43.0 | 11.000000 | 32.0 | 37.5 | 43.0 | 48.5 | 54.0 |
| Vw | 2.0 | 66.0 | 50.911688 | 30.0 | 48.0 | 66.0 | 84.0 | 102.0 |
We can also use several aggregators at once by aggregate
g["hp"].aggregate(["min", "max"])
| min | max | |
|---|---|---|
| brand | ||
| Mercedes | 50 | 50 |
| Tesla | 32 | 54 |
| Vw | 30 | 102 |
Pivoting#
Another way of getting overview of the data is by pivoting (somewhat like groupby in multiple “keys”).
# View "hp" just as a function of brand where the remaining dimensions are aggregated
data.pivot_table("hp", index="brand", aggfunc="sum")
| hp | |
|---|---|
| brand | |
| Mercedes | 50 |
| Tesla | 129 |
| Vw | 132 |
# View "hp" just as a function of brand and type (leaving agg on models). WE have 2 types SUV and SEDAN
data.pivot_table("hp", index="brand", columns=["type"], aggfunc="sum")
| type | sedan | suv |
|---|---|---|
| brand | ||
| Mercedes | NaN | 50.0 |
| Tesla | 75.0 | 54.0 |
| Vw | 30.0 | 102.0 |
# Filling in missing data
data.pivot_table("hp", index="brand", columns=["model"], aggfunc="sum")
| model | 3 | Gclass | S | X | passat | tuareg |
|---|---|---|---|---|---|---|
| brand | ||||||
| Mercedes | NaN | 50.0 | NaN | NaN | NaN | NaN |
| Tesla | 43.0 | NaN | 32.0 | 54.0 | NaN | NaN |
| Vw | NaN | NaN | NaN | NaN | 30.0 | 102.0 |
Hands on examples#
Let’s explore some datasets
1. Used cars
This dataset is downloaded from Kaggle and statistics on saled cars (ebay?). More precisely, having read the data by built-in
reader read_csv we have
from pathlib import Path
import requests
def download_file(filename, url):
path = Path(filename)
if path.exists():
return path
print(f"Downloading {path}")
with requests.get(url, stream=True) as r:
r.raise_for_status()
with path.open("wb") as f:
for chunk in r.iter_content(chunk_size=8192):
f.write(chunk)
return path
url = "https://www.kaggle.com/datasets/tsaustin/us-used-car-sales-data/download?datasetVersionNumber=4"
filename = Path("data") / "used_car_sales.csv"
# download_file(filename, url)
data = pd.read_csv(
"./data/used_car_sales.csv", dtype={"Mileage": int, "pricesold": int}
)
data.columns
Index(['ID', 'pricesold', 'yearsold', 'zipcode', 'Mileage', 'Make', 'Model',
'Year', 'Trim', 'Engine', 'BodyType', 'NumCylinders', 'DriveType'],
dtype='object')
data.head(10)
| ID | pricesold | yearsold | zipcode | Mileage | Make | Model | Year | Trim | Engine | BodyType | NumCylinders | DriveType | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 137178 | 7500 | 2020 | 786** | 84430 | Ford | Mustang | 1988 | LX | 5.0L Gas V8 | Sedan | 0 | RWD |
| 1 | 96705 | 15000 | 2019 | 81006 | 0 | Replica/Kit Makes | Jaguar Beck Lister | 1958 | NaN | 383 Fuel injected | Convertible | 8 | RWD |
| 2 | 119660 | 8750 | 2020 | 33449 | 55000 | Jaguar | XJS | 1995 | 2+2 Cabriolet | 4.0L In-Line 6 Cylinder | Convertible | 6 | RWD |
| 3 | 80773 | 11600 | 2019 | 07852 | 97200 | Ford | Mustang | 1968 | Stock | 289 cu. in. V8 | Coupe | 8 | RWD |
| 4 | 64287 | 44000 | 2019 | 07728 | 40703 | Porsche | 911 | 2002 | Turbo X-50 | 3.6L | Coupe | 6 | AWD |
| 5 | 132695 | 950 | 2020 | 462** | 71300 | Mercury | Montclair | 1965 | NaN | NO ENGINE | Sedan | 0 | RWD |
| 6 | 132829 | 950 | 2020 | 105** | 71300 | Mercury | Montclair | 1965 | NaN | NaN | Sedan | 0 | NaN |
| 7 | 5250 | 70000 | 2019 | 07627 | 6500 | Land Rover | Defender | 1997 | NaN | 4.0 Liter Fuel Injected V8 | NaN | 0 | 4WD |
| 8 | 29023 | 1330 | 2019 | 07043 | 167000 | Honda | Civic | 2001 | EX | NaN | Coupe | 4 | FWD |
| 9 | 80293 | 25200 | 2019 | 33759 | 15000 | Pontiac | GTO | 1970 | NaN | NaN | NaN | 0 | NaN |
As part of exploring the dataset let’s see about the distributiion of the mileage of the cars. We use cut to comnpute a “bin” for each item
max_distance = data["Mileage"].max()
# max_distance
mileage_dist = pd.cut(
data["Mileage"], bins=np.linspace(0, 1_000_000, 10, endpoint=True)
)
mileage_dist
0 (0.0, 111111.111]
1 NaN
2 (0.0, 111111.111]
3 (0.0, 111111.111]
4 (0.0, 111111.111]
...
122139 (0.0, 111111.111]
122140 (111111.111, 222222.222]
122141 (0.0, 111111.111]
122142 (111111.111, 222222.222]
122143 (111111.111, 222222.222]
Name: Mileage, Length: 122144, dtype: category
Categories (9, interval[float64, right]): [(0.0, 111111.111] < (111111.111, 222222.222] < (222222.222, 333333.333] < (333333.333, 444444.444] ... (555555.556, 666666.667] < (666666.667, 777777.778] < (777777.778, 888888.889] < (888888.889, 1000000.0]]
# Let's see about the distribution
mileage_dist.value_counts()
Mileage
(0.0, 111111.111] 73813
(111111.111, 222222.222] 37788
(222222.222, 333333.333] 4885
(888888.889, 1000000.0] 1112
(333333.333, 444444.444] 404
(444444.444, 555555.556] 73
(555555.556, 666666.667] 58
(666666.667, 777777.778] 52
(777777.778, 888888.889] 41
Name: count, dtype: int64
We can find out which manufacturer sold the most cars, by grouping and then collapsing based on size
data.groupby("Make").size()
Make
1964 International 1
2101 1
300 1
AC 1
AC Cobra 1
..
ponitac 1
rambler 1
renault 1
smart 1
willy's jeep 1
Length: 464, dtype: int64
# A sanity check
data.groupby("Make").size().sum() == len(data)
# Unpack this
data.groupby("Make").size().sort_values(ascending=False).head(10)
Make
Ford 22027
Chevrolet 21171
Toyota 6676
Mercedes-Benz 6241
Dodge 5899
BMW 5128
Jeep 4543
Cadillac 3657
Volkswagen 3589
Honda 3451
dtype: int64
data["Make"].value_counts().sort_values(ascending=False).head(10)
Make
Ford 22027
Chevrolet 21171
Toyota 6676
Mercedes-Benz 6241
Dodge 5899
BMW 5128
Jeep 4543
Cadillac 3657
Volkswagen 3589
Honda 3451
Name: count, dtype: int64
We might wonder how the price is related to the car attributes. Let’s consider it first as a function of milage
Question: Are these correlated?
fix, ax = plt.subplots()
# We take the mean in others
data.pivot_table("pricesold", index="Mileage", aggfunc="mean").plot(
ax=ax, marker="x", linestyle="none"
)
ax.set_xscale("log")
ax.set_yscale("log")
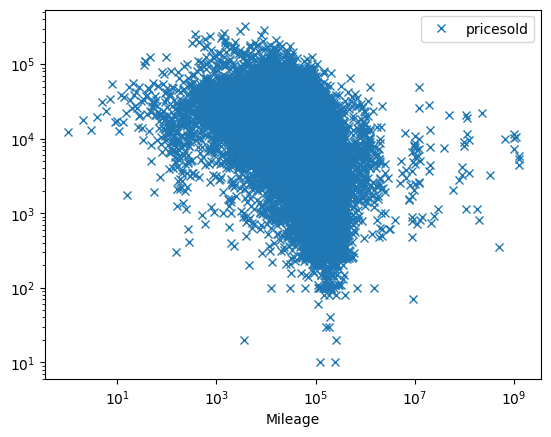
Does it vary with year?
fix, ax = plt.subplots(1, 3, sharey=True, figsize=(16, 10))
# We take the mean in others
tab = data.pivot_table(
"pricesold", index="Mileage", columns=["yearsold"], aggfunc="mean"
)
for i, year in enumerate((2018, 2019, 2020)):
tab[year].plot(ax=ax[i], marker="x", linestyle="none")
ax[i].set_xscale("log")
ax[i].set_yscale("log")
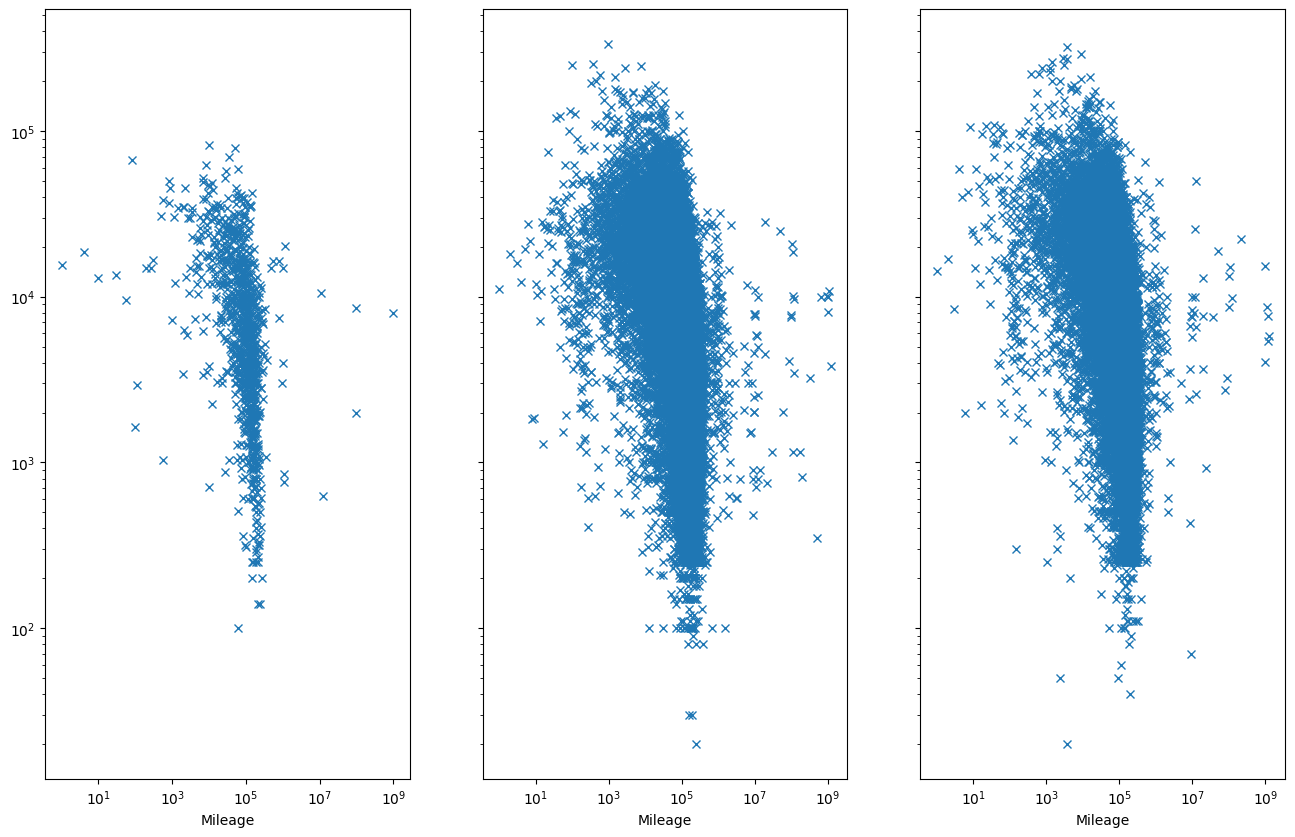
At this point we would like to see about the role of the cars age. However, we find out there’s something off with the data.
data["Year"].describe()
# We see that we have 92, 0, 20140000
count 1.221440e+05
mean 3.959362e+03
std 1.984514e+05
min 0.000000e+00
25% 1.977000e+03
50% 2.000000e+03
75% 2.008000e+03
max 2.014000e+07
Name: Year, dtype: float64
We show one way of dealing with strangely formated data in the next example. In particular, we will address the problem already when reading the data.
2. People in Norway by region and age in 2022
The dateset is obtained from SSB and we see that the encoding of the age will present some difficulty for numerics.
with open("./data/nor_population2022.csv") as f:
ln = 0
while ln < 5:
print(next(f).strip())
ln += 1
"region";"alder";"ar";"xxx";"count"
"30 Viken";"000 0 ar";"2022";"Personer";12620
"30 Viken";"001 1 ar";"2022";"Personer";12512
"30 Viken";"002 2 ar";"2022";"Personer";13203
"30 Viken";"003 3 ar";"2022";"Personer";13727
Our solution is to specify a converter for the column which uses regexp to get the age string which is converted to integer
import re
# And we also get rid of some redundant data
data = pd.read_csv(
"./data/nor_population2022.csv",
delimiter=";",
converters={"alder": lambda x: int(re.search(r"(\d+) (ar)", x).group(1))},
).drop(["ar", "xxx"], axis="columns")
data
| region | alder | count | |
|---|---|---|---|
| 0 | 30 Viken | 0 | 12620 |
| 1 | 30 Viken | 1 | 12512 |
| 2 | 30 Viken | 2 | 13203 |
| 3 | 30 Viken | 3 | 13727 |
| 4 | 30 Viken | 4 | 14003 |
| ... | ... | ... | ... |
| 1267 | 21 Svalbard | 101 | 0 |
| 1268 | 21 Svalbard | 102 | 0 |
| 1269 | 21 Svalbard | 103 | 0 |
| 1270 | 21 Svalbard | 104 | 0 |
| 1271 | 21 Svalbard | 105 | 0 |
1272 rows × 3 columns
Question: Can you now fix the problem with car dataset?
We can have a look at the populaton in different regions
data.groupby("region")["count"].sum().plot()
plt.xticks(rotation=45);
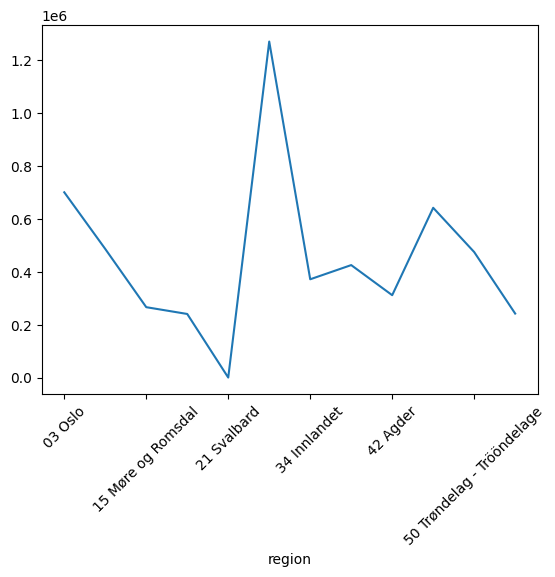
Or breakdown the distribution
fig, ax = plt.subplots()
data.pivot_table("count", index="alder", columns=["region"]).plot(ax=ax)
ax.legend(bbox_to_anchor=(1.05, 0.5))
<matplotlib.legend.Legend at 0x7f8a493da880>
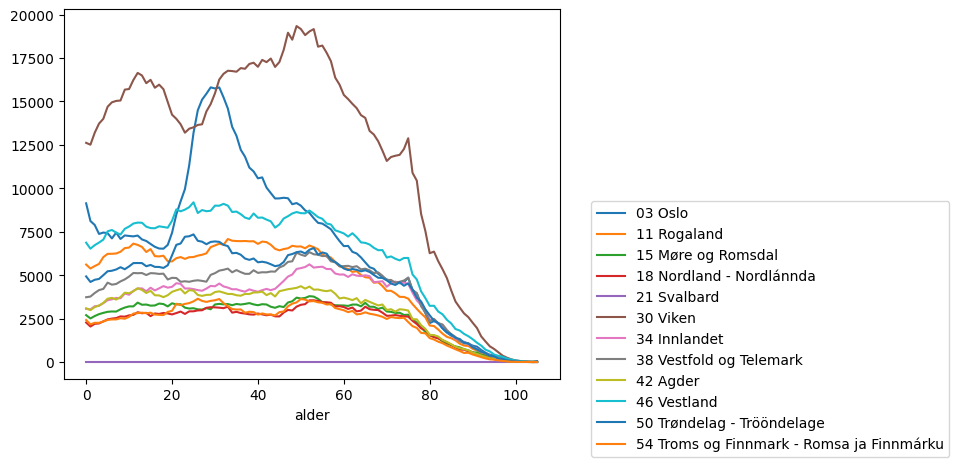
3. Joining
This example is adopted from * Python Data Science Handbook by Jake VanderPlas. We first download the datasets
for name in "abbrevs", "areas", "population":
fname = f"state-{name}.csv"
download_file(
f"data/{fname}",
url=f"https://raw.githubusercontent.com/jakevdp/data-USstates/HEAD/{fname}",
)
You can see that they contain data [full name -> population], [full_name -> area], [abbreviation -> full_name]. Given this we want to order the stated by population density
abbrevs = pd.read_csv("./data/state-abbrevs.csv")
areas = pd.read_csv("./data/state-areas.csv")
pop = pd.read_csv("./data/state-population.csv")
abbrevs.head(10)
| state | abbreviation | |
|---|---|---|
| 0 | Alabama | AL |
| 1 | Alaska | AK |
| 2 | Arizona | AZ |
| 3 | Arkansas | AR |
| 4 | California | CA |
| 5 | Colorado | CO |
| 6 | Connecticut | CT |
| 7 | Delaware | DE |
| 8 | District of Columbia | DC |
| 9 | Florida | FL |
(areas.columns, pop.columns, abbrevs.columns)
(Index(['state', 'area (sq. mi)'], dtype='object'),
Index(['state/region', 'ages', 'year', 'population'], dtype='object'),
Index(['state', 'abbreviation'], dtype='object'))
We want to build a table which has both areas and population info. To get there population should get abbreviation so that we can look up areas.
merged = pd.merge(
pop, abbrevs, left_on="state/region", right_on="abbreviation", how="outer"
).drop("abbreviation", axis=1)
merged.head(5)
| state/region | ages | year | population | state | |
|---|---|---|---|---|---|
| 0 | AL | under18 | 2012 | 1117489.0 | Alabama |
| 1 | AL | total | 2012 | 4817528.0 | Alabama |
| 2 | AL | under18 | 2010 | 1130966.0 | Alabama |
| 3 | AL | total | 2010 | 4785570.0 | Alabama |
| 4 | AL | under18 | 2011 | 1125763.0 | Alabama |
We inspect the integrity of dataset. Something is wrong with state/region which is a problem if we want to map further.
merged.isnull().any()
state/region False
ages False
year False
population True
state True
dtype: bool
By further inspection the missing data is for “not the usual 50 states” and so we just drop them
merged.loc[merged["state"].isnull(), "state/region"].unique()
array(['PR', 'USA'], dtype=object)
merged.dropna(inplace=True)
We can add to our table (left merge) the areas
final = pd.merge(merged, areas, on="state")
final.head(5)
| state/region | ages | year | population | state | area (sq. mi) | |
|---|---|---|---|---|---|---|
| 0 | AL | under18 | 2012 | 1117489.0 | Alabama | 52423 |
| 1 | AL | total | 2012 | 4817528.0 | Alabama | 52423 |
| 2 | AL | under18 | 2010 | 1130966.0 | Alabama | 52423 |
| 3 | AL | total | 2010 | 4785570.0 | Alabama | 52423 |
| 4 | AL | under18 | 2011 | 1125763.0 | Alabama | 52423 |
We use query (SQL like) language to get the right rows
table = final.query(
"year == 2012 & ages == 'total'"
) # final['population']/final['area (sq. mi)']
table.head(10)
| state/region | ages | year | population | state | area (sq. mi) | |
|---|---|---|---|---|---|---|
| 1 | AL | total | 2012 | 4817528.0 | Alabama | 52423 |
| 95 | AK | total | 2012 | 730307.0 | Alaska | 656425 |
| 97 | AZ | total | 2012 | 6551149.0 | Arizona | 114006 |
| 191 | AR | total | 2012 | 2949828.0 | Arkansas | 53182 |
| 193 | CA | total | 2012 | 37999878.0 | California | 163707 |
| 287 | CO | total | 2012 | 5189458.0 | Colorado | 104100 |
| 289 | CT | total | 2012 | 3591765.0 | Connecticut | 5544 |
| 383 | DE | total | 2012 | 917053.0 | Delaware | 1954 |
| 385 | DC | total | 2012 | 633427.0 | District of Columbia | 68 |
| 479 | FL | total | 2012 | 19320749.0 | Florida | 65758 |
We will use the states to index into the column. By index preservation this will give us states to density
table.set_index("state", inplace=True)
density = table["population"] / table["area (sq. mi)"]
density.head(5)
state
Alabama 91.897221
Alaska 1.112552
Arizona 57.463195
Arkansas 55.466662
California 232.121278
dtype: float64
density.sort_values(ascending=False).head(10)
state
District of Columbia 9315.102941
New Jersey 1016.710502
Rhode Island 679.808414
Connecticut 647.865260
Massachusetts 629.588157
Maryland 474.318369
Delaware 469.320880
New York 359.359798
Florida 293.815946
Pennsylvania 277.139151
dtype: float64
Question: Similar data can be obtained from Eurostat. The new thing is that the format .tsv. Let’s start parsing the population data
with open("./data/eustat_population.tsv") as f:
ln = 0
while ln < 10:
print(next(f).strip())
ln += 1
unit,age,sex,geo\time 2010 2011 2012 2013 2014 2015 2016 2017 2018 2019 2020 2021
NR,TOTAL,T,AL01 : : : : : : 848059 826904 819793 813758 804689 797955
NR,TOTAL,T,AL02 : : : : : : 1110562 1146183 1162544 1170142 1176240 1178435
NR,TOTAL,T,AL03 : : : : : : 927405 903504 887987 878527 865026 853351
NR,TOTAL,T,ALXX : : : : : : : : 0 0 : :
NR,TOTAL,T,AT11 283697 284581 285782 286691 287416 288356 291011 291942 292675 293433 294436 296010
NR,TOTAL,T,AT12 1605897 1609474 1614455 1618592 1625485 1636778 1653691 1665753 1670668 1677542 1684287 1690879
NR,TOTAL,T,AT13 1689995 1702855 1717084 1741246 1766746 1797337 1840226 1867582 1888776 1897491 1911191 1920949
NR,TOTAL,T,AT21 557998 556718 556027 555473 555881 557641 560482 561077 560898 560939 561293 562089
NR,TOTAL,T,AT22 1205045 1206611 1208696 1210971 1215246 1221570 1232012 1237298 1240214 1243052 1246395 1247077
We see that the countries are subdivided. Let’s figure out the reader to help here
d = pd.read_csv(
"./data/eustat_population.tsv",
sep="\t",
converters={0: lambda x: re.search(r"([A-Z]{2})(\w\w)$", x).group(1)},
)
d
| unit,age,sex,geo\time | 2010 | 2011 | 2012 | 2013 | 2014 | 2015 | 2016 | 2017 | 2018 | 2019 | 2020 | 2021 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | AL | : | : | : | : | : | : | 848059 | 826904 | 819793 | 813758 | 804689 | 797955 |
| 1 | AL | : | : | : | : | : | : | 1110562 | 1146183 | 1162544 | 1170142 | 1176240 | 1178435 |
| 2 | AL | : | : | : | : | : | : | 927405 | 903504 | 887987 | 878527 | 865026 | 853351 |
| 3 | AL | : | : | : | : | : | : | : | : | 0 | 0 | : | : |
| 4 | AT | 283697 | 284581 | 285782 | 286691 | 287416 | 288356 | 291011 | 291942 | 292675 | 293433 | 294436 | 296010 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 337 | UK | 463388 | 465794 | 466605 | 466737 | 467326 | 467889 | 468903 | 469420 | 470743 | 470990 | : | : |
| 338 | UK | : | : | 1915517 | 1924614 | 1933992 | 1946564 | 1961928 | 1976392 | 1988307 | 1998699 | : | : |
| 339 | UK | : | : | 1499887 | 1500987 | 1504303 | 1510438 | 1520628 | 1531216 | 1536415 | 1541998 | : | : |
| 340 | UK | : | : | 946492 | 945666 | 944842 | 944507 | 945233 | 946372 | 946837 | 947186 | : | : |
| 341 | UK | 1799019 | 1809539 | 1818995 | 1826761 | 1835290 | 1846229 | 1857048 | 1866638 | 1875957 | : | : | : |
342 rows × 13 columns
The next challenge is to handle the missing values which do not show with d.isna() as all the values are strings and the missing is “: “. How would you solve this issue?
4. Timeseries analysis
Our final examples follows Chapter 4. in PDSH. We will be looking at statics from sensors counting bikers on a bridge. We have 2 counters one for left/right side each.
download_file(
"data/FremontBridge.csv",
url="https://data.seattle.gov/api/views/65db-xm6k/rows.csv?accessType=DOWNLOAD",
)
data = pd.read_csv("data/FremontBridge.csv", index_col="Date", parse_dates=True)
data.head()
/tmp/ipykernel_419026/796841783.py:5: UserWarning: Could not infer format, so each element will be parsed individually, falling back to `dateutil`. To ensure parsing is consistent and as-expected, please specify a format.
data = pd.read_csv("data/FremontBridge.csv", index_col="Date", parse_dates=True)
| Fremont Bridge Sidewalks, south of N 34th St | Fremont Bridge Sidewalks, south of N 34th St Cyclist East Sidewalk | Fremont Bridge Sidewalks, south of N 34th St Cyclist West Sidewalk | |
|---|---|---|---|
| Date | |||
| 2022-08-01 00:00:00 | 23.0 | 7.0 | 16.0 |
| 2022-08-01 01:00:00 | 12.0 | 5.0 | 7.0 |
| 2022-08-01 02:00:00 | 3.0 | 0.0 | 3.0 |
| 2022-08-01 03:00:00 | 5.0 | 2.0 | 3.0 |
| 2022-08-01 04:00:00 | 10.0 | 2.0 | 8.0 |
len(data)
95640
For the next steps we will only consider the total count of bikers so we simplify the table
data.columns = ["Total", "East", "West"]
data.drop("East", axis="columns", inplace=True)
data.drop("West", axis="columns", inplace=True)
data
| Total | |
|---|---|
| Date | |
| 2022-08-01 00:00:00 | 23.0 |
| 2022-08-01 01:00:00 | 12.0 |
| 2022-08-01 02:00:00 | 3.0 |
| 2022-08-01 03:00:00 | 5.0 |
| 2022-08-01 04:00:00 | 10.0 |
| ... | ... |
| 2023-08-31 19:00:00 | 224.0 |
| 2023-08-31 20:00:00 | 142.0 |
| 2023-08-31 21:00:00 | 67.0 |
| 2023-08-31 22:00:00 | 43.0 |
| 2023-08-31 23:00:00 | 12.0 |
95640 rows × 1 columns
Marking at the data we can definitely see Covid19 but also some seasonal variations
data.plot()
plt.ylabel("Hourly Bicycle Count"); # NOTE: that hour data is for every hour
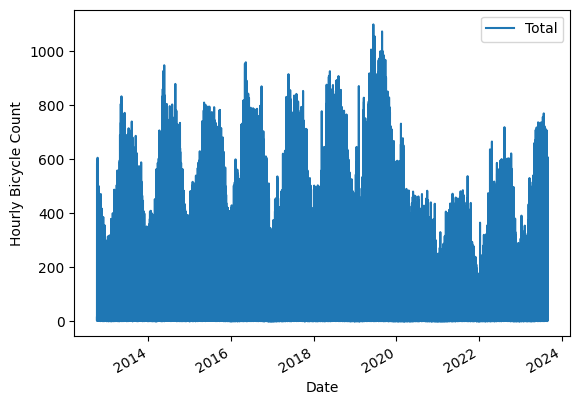
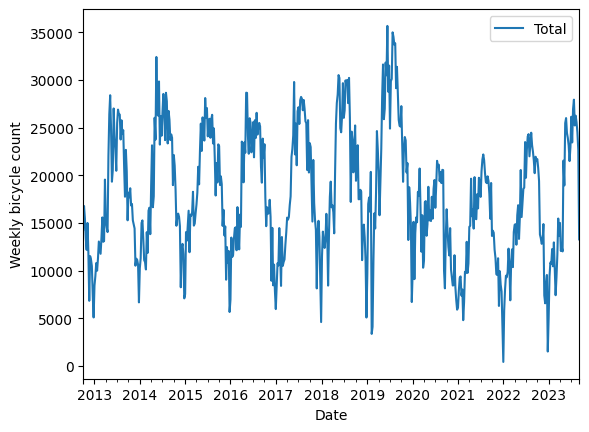
We can downsample the signal to reveal the variations within year better. Below we downsample to Weeks
weekly = data.resample("W").sum()
# Confirmation
weekly.index
DatetimeIndex(['2012-10-07', '2012-10-14', '2012-10-21', '2012-10-28',
'2012-11-04', '2012-11-11', '2012-11-18', '2012-11-25',
'2012-12-02', '2012-12-09',
...
'2023-07-02', '2023-07-09', '2023-07-16', '2023-07-23',
'2023-07-30', '2023-08-06', '2023-08-13', '2023-08-20',
'2023-08-27', '2023-09-03'],
dtype='datetime64[ns]', name='Date', length=570, freq='W-SUN')
# And again
weekly.index.isocalendar().week
Date
2012-10-07 40
2012-10-14 41
2012-10-21 42
2012-10-28 43
2012-11-04 44
..
2023-08-06 31
2023-08-13 32
2023-08-20 33
2023-08-27 34
2023-09-03 35
Freq: W-SUN, Name: week, Length: 570, dtype: UInt32
weekly.plot(style=["-"])
plt.ylabel("Weekly bicycle count");
Recall that having time series as index enables many convience functions. For example, we can group/bin data time points by hour and reduce on the subindices giving hous an hourly mean bike usage. It appears to correlate well with rush hours.
by_time = data.groupby(data.index.time).mean()
hourly_ticks = 4 * 60 * 60 * np.arange(6) # 3600 * 24, every 4 hour
by_time.plot(xticks=hourly_ticks, style=["-"]);
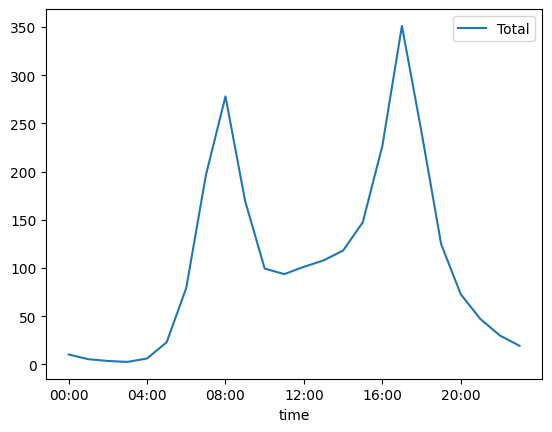
We can repeat the same exercise, this time breaking up the data by hours to reveal that bikes are probably most used for commuting to work
by_weekday = data.groupby(data.index.dayofweek).mean()
by_weekday.index = ["Mon", "Tues", "Wed", "Thurs", "Fri", "Sat", "Sun"]
by_weekday.plot(style=["-"]);
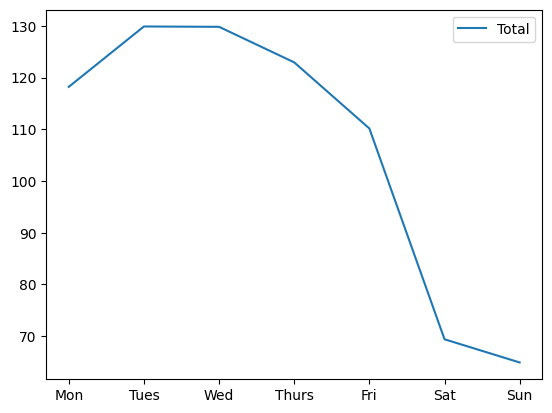
Another look at the same thing using pivot_table
import matplotlib.colors as colors
Z = data.pivot_table(
"Total", index=data.index.weekday, columns=[data.index.time]
).values
plt.pcolor(Z, norm=colors.LogNorm(vmin=Z.min(), vmax=Z.max()), cmap="inferno")
plt.colorbar()
<matplotlib.colorbar.Colorbar at 0x7f8a46820460>
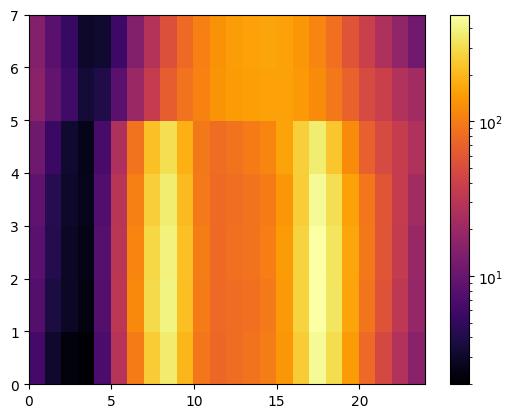
The heatmap suggests that there are 2 peaks in weekdays and a single one during the weekend. We can varify this by essentially collapsing/averaging the plot in the vertical direction separately for the two categories of days. Let’s compute the mask for each day in the series
weekend = np.where(data.index.weekday < 5, "Weekday", "Weekend")
Using the mask we can group the data first by the day catogory and then by hours
by_time = data.groupby([weekend, data.index.time]).mean()
by_time.index[:4]
MultiIndex([('Weekday', 00:00:00),
('Weekday', 01:00:00),
('Weekday', 02:00:00),
('Weekday', 03:00:00)],
)
Finally we have the two collapsed plots
import matplotlib.pyplot as plt
fig, ax = plt.subplots(1, 2, figsize=(14, 5))
by_time.loc["Weekday"].plot(
ax=ax[0], title="Weekdays", xticks=hourly_ticks, style=["-"]
)
by_time.loc["Weekend"].plot(
ax=ax[1], title="Weekends", xticks=hourly_ticks, style=["-"], color="red"
);
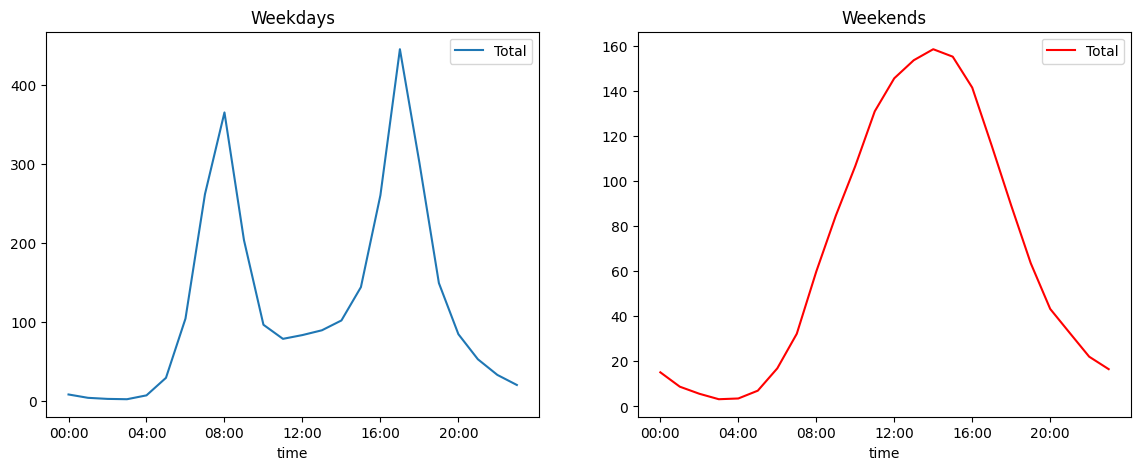
Question: Head on to Oslo Bysykkel web and get the trip data (as CSV). In the last years the students analyzed the trips by distance (see the IN3110 course webpage). What about the distribution of trips by the average speed? Can you use this to infer if the trip was going (on average) uphill or downhill?
bike_csv = "./data/oslo_bike_2022_09.csv"
download_file(
bike_csv, url="https://data.urbansharing.com/oslobysykkel.no/trips/v1/2022/09.csv"
)
PosixPath('data/oslo_bike_2022_09.csv')
with open(bike_csv) as f:
print(next(f).strip())
print(next(f).strip())
started_at,ended_at,duration,start_station_id,start_station_name,start_station_description,start_station_latitude,start_station_longitude,end_station_id,end_station_name,end_station_description,end_station_latitude,end_station_longitude
2022-09-01 03:04:31.178000+00:00,2022-09-01 03:13:01.298000+00:00,510,437,Sentrum Scene,ved Arbeidersamfunnets plass,59.91546786564256,10.751140873016311,583,Galgeberg,langs St. Halvards gate,59.90707579234818,10.779164402446728
At this point we know that time info should be parsed with dates. For duration (in seconds) we will have ints
data = pd.read_csv(
bike_csv,
parse_dates=["started_at", "ended_at"],
dtype={"duration": int},
)
data.head(5)
| started_at | ended_at | duration | start_station_id | start_station_name | start_station_description | start_station_latitude | start_station_longitude | end_station_id | end_station_name | end_station_description | end_station_latitude | end_station_longitude | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 2022-09-01 03:04:31.178000+00:00 | 2022-09-01 03:13:01.298000+00:00 | 510 | 437 | Sentrum Scene | ved Arbeidersamfunnets plass | 59.915468 | 10.751141 | 583 | Galgeberg | langs St. Halvards gate | 59.907076 | 10.779164 |
| 1 | 2022-09-01 03:11:09.104000+00:00 | 2022-09-01 03:14:52.506000+00:00 | 223 | 578 | Hallings gate | langs Dalsbergstien | 59.922777 | 10.738655 | 499 | Bjerregaards gate | ovenfor Fredrikke Qvams gate | 59.925488 | 10.746058 |
| 2 | 2022-09-01 03:11:37.861000+00:00 | 2022-09-01 03:23:23.939000+00:00 | 706 | 421 | Alexander Kiellands Plass | langs Maridalsveien | 59.928067 | 10.751203 | 390 | Saga Kino | langs Olav Vs gate | 59.914240 | 10.732771 |
| 3 | 2022-09-01 03:13:00.843000+00:00 | 2022-09-01 03:17:17.639000+00:00 | 256 | 735 | Oslo Hospital | ved trikkestoppet | 59.903213 | 10.767344 | 465 | Bjørvika | under broen Nylandsveien | 59.909006 | 10.756180 |
| 4 | 2022-09-01 03:13:13.330000+00:00 | 2022-09-01 03:24:15.758000+00:00 | 662 | 525 | Myraløkka Øst | ved Bentsenbrua | 59.937205 | 10.760581 | 443 | Sjøsiden ved trappen | Oslo S | 59.910154 | 10.751981 |
What is the station with most departures?
data["start_station_id"].value_counts().sort_values(ascending=False)
start_station_id
421 2133
1755 2079
480 1984
398 1970
551 1918
...
591 84
1919 74
454 72
560 58
573 44
Name: count, Length: 263, dtype: int64
We will approximate the trip distance by measing it as the geodesic between start and end points.
from math import asin, cos, pi, sin, sqrt
def trip_distance(row):
"""As the crow flies"""
lat_station = row["start_station_latitude"]
lon_station = row["start_station_longitude"]
lat_sentrum = row["end_station_latitude"]
lon_sentrum = row["end_station_longitude"]
degrees = pi / 180 # convert degrees to radians
a = (
0.5
- (cos((lat_sentrum - lat_station) * degrees) / 2)
+ (
cos(lat_sentrum * degrees)
* cos(lat_station * degrees)
* (1 - cos((lon_station - lon_sentrum) * degrees))
/ 2
)
)
# We have the return value in meters
return 12742_000 * asin(sqrt(a)) # 2 * R * asin...
distance = data.apply(trip_distance, axis="columns")
duration = data["duration"]
speed = distance / duration
speed.hist()
<AxesSubplot: >
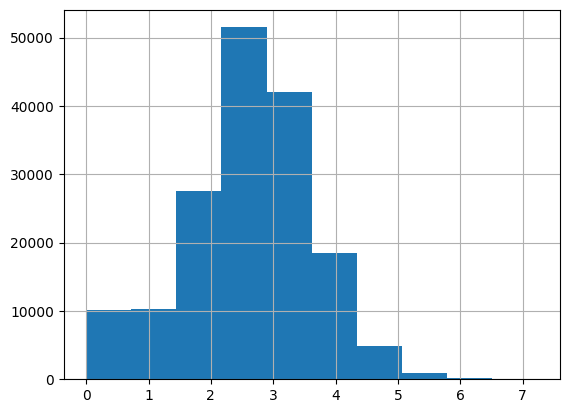
Bonus: Visualization
from ipyleaflet import Circle, Map, Marker, Polyline, basemap_to_tiles, basemaps
from ipywidgets import HTML
trip_csv = "./data/oslo_bike_2022_09.csv"
trips = pd.read_csv(trip_csv)
station_data = trips.groupby(
[
"start_station_id",
"start_station_longitude",
"start_station_latitude",
"start_station_name",
]
).count()
station_data = station_data.reset_index()
station_data = station_data.drop(columns=station_data.columns[-7:])
station_data = station_data.rename(columns={"started_at": "started_trips"})
station_data = station_data.set_index("start_station_id")
station_data["ended_trips"] = trips["end_station_id"].value_counts()
oslo_center = (
59.9127,
10.7461,
) # NB ipyleaflet uses Lat-Long (i.e. y,x, when specifying coordinates)
oslo_map = Map(center=oslo_center, zoom=13, close_popup_on_click=False)
def add_markers(row):
center = row["start_station_latitude"], row["start_station_longitude"]
marker = Marker(location=center)
marker2 = Circle(
location=center, radius=int(0.04 * row["ended_trips"]), color="red"
)
message = HTML()
message.value = (
f"{row['start_station_name']} <br> Trips started: {row['started_trips']}"
)
marker.popup = message
oslo_map.add_layer(marker)
oslo_map.add_layer(marker2)
station_data.apply(add_markers, axis=1)
start_station_id
377 None
378 None
380 None
381 None
382 None
...
2340 None
2347 None
2349 None
2350 None
2351 None
Length: 263, dtype: object
oslo_map